The Copper Efflux Regulator CueR Is Subject to ATP-Dependent Proteolysis in Escherichia coli
- PMID: 28293558
- PMCID: PMC5329002
- DOI: 10.3389/fmolb.2017.00009
The Copper Efflux Regulator CueR Is Subject to ATP-Dependent Proteolysis in Escherichia coli
Abstract
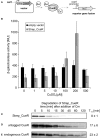
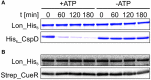
The trace element copper serves as cofactor for many enzymes but is toxic at elevated concentrations. In bacteria, the intracellular copper level is maintained by copper efflux systems including the Cue system controlled by the transcription factor CueR. CueR, a member of the MerR family, forms homodimers, and binds monovalent copper ions with high affinity. It activates transcription of the copper tolerance genes copA and cueO via a conserved DNA-distortion mechanism. The mechanism how CueR-induced transcription is turned off is not fully understood. Here, we report that Escherichia coli CueR is prone to proteolysis by the AAA+ proteases Lon, ClpXP, and ClpAP. Using a set of CueR variants, we show that CueR degradation is not altered by mutations affecting copper binding, dimerization or DNA binding of CueR, but requires an accessible C terminus. Except for a twofold stabilization shortly after a copper pulse, proteolysis of CueR is largely copper-independent. Our results suggest that ATP-dependent proteolysis contributes to copper homeostasis in E. coli by turnover of CueR, probably to allow steady monitoring of changes of the intracellular copper level and shut-off of CueR-dependent transcription.
Keywords: AAA+ proteases; ClpAP; ClpXP; CueR; Lon; MerR family; copper homoeostasis; proteolysis.
Figures





Similar articles
-
CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA.Mol Microbiol. 2001 Jan;39(2):502-11. doi: 10.1046/j.1365-2958.2001.02264.x. Mol Microbiol. 2001. PMID: 11136469
-
The Copper Efflux Regulator (CueR).Subcell Biochem. 2024;104:17-31. doi: 10.1007/978-3-031-58843-3_2. Subcell Biochem. 2024. PMID: 38963481 Review.
-
Structural basis of copper-efflux-regulator-dependent transcription activation.iScience. 2021 Apr 16;24(5):102449. doi: 10.1016/j.isci.2021.102449. eCollection 2021 May 21. iScience. 2021. PMID: 34113812 Free PMC article.
-
Transcriptional response of Escherichia coli to external copper.Mol Microbiol. 2005 Apr;56(1):215-27. doi: 10.1111/j.1365-2958.2005.04532.x. Mol Microbiol. 2005. PMID: 15773991
-
Copper-responsive gene regulation in bacteria.Microbiology (Reading). 2012 Oct;158(Pt 10):2451-2464. doi: 10.1099/mic.0.058487-0. Epub 2012 Aug 23. Microbiology (Reading). 2012. PMID: 22918892 Review.
Cited by
-
The ropAe gene encodes a porin-like protein involved in copper transit in Rhizobium etli CFN42.Microbiologyopen. 2018 Jun;7(3):e00573. doi: 10.1002/mbo3.573. Epub 2017 Dec 27. Microbiologyopen. 2018. PMID: 29280343 Free PMC article.
-
A genome-wide screen reveals the involvement of enterobactin-mediated iron acquisition in Escherichia coli survival during copper stress.Metallomics. 2021 Sep 6;13(9):mfab052. doi: 10.1093/mtomcs/mfab052. Metallomics. 2021. PMID: 34415046 Free PMC article.
-
Metabolic Sensing of Extracytoplasmic Copper Availability via Translational Control by a Nascent Exported Protein.mBio. 2023 Feb 28;14(1):e0304022. doi: 10.1128/mbio.03040-22. Epub 2023 Jan 4. mBio. 2023. PMID: 36598193 Free PMC article.
-
U-shaped nonlinear relationship between dietary copper intake and peripheral neuropathy.Sci Rep. 2024 Oct 25;14(1):25263. doi: 10.1038/s41598-024-76159-6. Sci Rep. 2024. PMID: 39448725 Free PMC article.
-
Linking Copper-Associated Signal Transduction Systems with Their Environment in Marine Bacteria.Microorganisms. 2023 Apr 13;11(4):1012. doi: 10.3390/microorganisms11041012. Microorganisms. 2023. PMID: 37110435 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

