Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling
- PMID: 28285822
- PMCID: PMC5376383
- DOI: 10.1016/j.neuron.2017.02.030
Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling
Abstract
The suprachiasmatic nucleus (SCN) of the hypothalamus orchestrates daily rhythms of physiology and behavior in mammals. Its circadian (∼24 hr) oscillations of gene expression and electrical activity are generated intrinsically and can persist indefinitely in temporal isolation. This robust and resilient timekeeping is generally regarded as a product of the intrinsic connectivity of its neurons. Here we show that neurons constitute only one "half" of the SCN clock, the one metabolically active during circadian daytime. In contrast, SCN astrocytes are active during circadian nighttime, when they suppress the activity of SCN neurons by regulating extracellular glutamate levels. This glutamatergic gliotransmission is sensed by neurons of the dorsal SCN via specific pre-synaptic NMDA receptor assemblies containing NR2C subunits. Remarkably, somatic genetic re-programming of intracellular clocks in SCN astrocytes was capable of remodeling circadian behavioral rhythms in adult mice. Thus, SCN circuit-level timekeeping arises from interdependent and mutually supportive astrocytic-neuronal signaling.
Keywords: GABA; NMDAR2C; SCN; astrocytic-neuronal interactions; calcium oscillations; circadian; circadian behavior; circuit synchronization; extracellular glutamate; membrane potential oscillations.
Copyright © 2017 MRC Laboratory of Molecular Biology. Published by Elsevier Inc. All rights reserved.
Figures
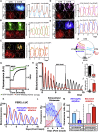
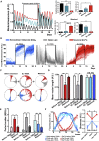
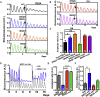
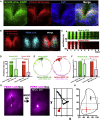

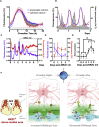

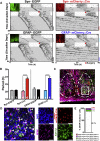
Comment in
-
Circadian rhythms: Astrocytes keep time.Nat Rev Neurosci. 2017 May;18(5):264. doi: 10.1038/nrn.2017.43. Epub 2017 Mar 31. Nat Rev Neurosci. 2017. PMID: 28360417 No abstract available.
Similar articles
-
Astrocytic Modulation of Neuronal Activity in the Suprachiasmatic Nucleus: Insights from Mathematical Modeling.J Biol Rhythms. 2020 Jun;35(3):287-301. doi: 10.1177/0748730420913672. Epub 2020 Apr 14. J Biol Rhythms. 2020. PMID: 32285754 Free PMC article.
-
Temporally chimeric mice reveal flexibility of circadian period-setting in the suprachiasmatic nucleus.Proc Natl Acad Sci U S A. 2016 Mar 29;113(13):3657-62. doi: 10.1073/pnas.1511351113. Epub 2016 Mar 10. Proc Natl Acad Sci U S A. 2016. PMID: 26966234 Free PMC article.
-
Cell-autonomous clock of astrocytes drives circadian behavior in mammals.Science. 2019 Jan 11;363(6423):187-192. doi: 10.1126/science.aat4104. Science. 2019. PMID: 30630934 Free PMC article.
-
Generation of circadian rhythms in the suprachiasmatic nucleus.Nat Rev Neurosci. 2018 Aug;19(8):453-469. doi: 10.1038/s41583-018-0026-z. Nat Rev Neurosci. 2018. PMID: 29934559 Review.
-
Collective timekeeping among cells of the master circadian clock.J Endocrinol. 2016 Jul;230(1):R27-49. doi: 10.1530/JOE-16-0054. Epub 2016 May 6. J Endocrinol. 2016. PMID: 27154335 Free PMC article. Review.
Cited by
-
Hyperammonaemia disrupts daily rhythms reversibly by elevating glutamate in the central circadian pacemaker.Liver Int. 2023 Mar;43(3):673-683. doi: 10.1111/liv.15476. Epub 2022 Nov 21. Liver Int. 2023. PMID: 36367321 Free PMC article.
-
Optogenetic targeting of cortical astrocytes selectively improves NREM sleep in an Alzheimer's disease mouse model.Sci Rep. 2024 Oct 4;14(1):23044. doi: 10.1038/s41598-024-73082-8. Sci Rep. 2024. PMID: 39362954 Free PMC article.
-
The Effect of Traumatic Brain Injury on Sleep Architecture and Circadian Rhythms in Mice-A Comparison of High-Frequency Head Impact and Controlled Cortical Injury.Biology (Basel). 2022 Jul 8;11(7):1031. doi: 10.3390/biology11071031. Biology (Basel). 2022. PMID: 36101412 Free PMC article.
-
A new player in circadian networks: Role of electrical synapses in regulating functions of the circadian clock.Front Physiol. 2022 Nov 3;13:968574. doi: 10.3389/fphys.2022.968574. eCollection 2022. Front Physiol. 2022. PMID: 36406999 Free PMC article. Review.
-
Insights about circadian clock in glioma: From molecular pathways to therapeutic drugs.CNS Neurosci Ther. 2022 Dec;28(12):1930-1941. doi: 10.1111/cns.13966. Epub 2022 Sep 6. CNS Neurosci Ther. 2022. PMID: 36066207 Free PMC article. Review.
References
-
- Abrahamson E.E., Moore R.Y. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. - PubMed
-
- Acker T.M., Yuan H., Hansen K.B., Vance K.M., Ogden K.K., Jensen H.S., Burger P.B., Mullasseril P., Snyder J.P., Liotta D.C., Traynelis S.F. Mechanism for noncompetitive inhibition by novel GluN2C/D N-methyl-D-aspartate receptor subunit-selective modulators. Mol. Pharmacol. 2011;80:782–795. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

