The activity of the carbamoyl phosphate synthase 1 promoter in human liver-derived cells is dependent on hepatocyte nuclear factor 3-beta
- PMID: 28272778
- PMCID: PMC5571533
- DOI: 10.1111/jcmm.13123
The activity of the carbamoyl phosphate synthase 1 promoter in human liver-derived cells is dependent on hepatocyte nuclear factor 3-beta
Abstract
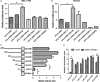
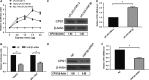
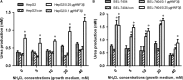
Carbamoyl phosphate synthase 1 (CPS1) is the rate-limiting enzyme in the first step of the urea cycle and an indispensable enzyme in the metabolism of human liver. However, CPS1 epigenetic regulation involves promoter analysis and the role of liver-enriched transcription factors (LETFs), which is not fully elucidated. In this work, the promoter region of hCPS1 gene was cloned, and its activity was investigated. An LETF, hepatocyte nuclear factor 3-beta (HNF3β), was found to promote the transcriptional expression of CPS1 in liver-derived cell lines. In addition, dual-luciferase reporter assay shows that the essential binding sites of the HNF3β may exist in the oligonucleotide -70 nt to +73 nt. Two putative binding sites are available for HNF3β. Mutation analysis results show that the binding site 2 of HNF3β was effective, and the transcriptional activity of CPS1 promoter significantly decreased after mutation. Electrophoretic mobile shift assay (EMSA) and ChIP assay confirmed that HNF3β can interact with the binding site in the CPS1 promoter region of -70 nt to +73 nt promoter region in vivo and in vitro to regulate the transcription of CPS1. Moreover, HNF3β overexpression enhanced the transcription of CPS1 and consequently improved the mRNA and protein levels of CPS1, whereas the knockdown of HNF3β showed the opposite effects. Finally, urea production in cells was measured, and ammonia detoxification improved significantly in cells after transfection with HNF3β. HNF3β plays a vital role in regulation of CPS1 gene and could promote the metabolism of ammonia by regulating CPS1 expression.
Keywords: ammonia detoxification; carbamoyl phosphate synthetase 1; hepatocyte nuclear factor 3-beta; liver-derived cell; promoter.
© 2017 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Figures

Similar articles
-
CPS1: Looking at an ancient enzyme in a modern light.Mol Genet Metab. 2020 Nov;131(3):289-298. doi: 10.1016/j.ymgme.2020.10.003. Epub 2020 Oct 10. Mol Genet Metab. 2020. PMID: 33317798 Free PMC article. Review.
-
Human liver fatty acid binding protein (hFABP1) gene is regulated by liver-enriched transcription factors HNF3β and C/EBPα.Biochimie. 2012 Feb;94(2):384-92. doi: 10.1016/j.biochi.2011.08.006. Epub 2011 Aug 16. Biochimie. 2012. PMID: 21856370
-
Generation of carbamoyl phosphate synthetase 1 reporter cell lines for the assessment of ammonia metabolism.J Cell Mol Med. 2017 Dec;21(12):3214-3223. doi: 10.1111/jcmm.13225. Epub 2017 May 30. J Cell Mol Med. 2017. PMID: 28557353 Free PMC article.
-
Y-box binding protein-1 down-regulates expression of carbamoyl phosphate synthetase-I by suppressing CCAAT enhancer-binding protein-alpha function in mice.Gastroenterology. 2009 Jul;137(1):330-40. doi: 10.1053/j.gastro.2009.02.064. Epub 2009 Mar 9. Gastroenterology. 2009. PMID: 19272383
-
Genetic, structural and biochemical basis of carbamoyl phosphate synthetase 1 deficiency.Mol Genet Metab. 2010 Dec;101(4):311-23. doi: 10.1016/j.ymgme.2010.08.002. Epub 2010 Aug 6. Mol Genet Metab. 2010. PMID: 20800523 Review.
Cited by
-
CRISPR/Cas9-based liver-derived reporter cells for screening of mPGES-1 inhibitors.J Enzyme Inhib Med Chem. 2019 Dec;34(1):799-807. doi: 10.1080/14756366.2019.1587416. J Enzyme Inhib Med Chem. 2019. PMID: 30879343 Free PMC article.
-
CPS1: Looking at an ancient enzyme in a modern light.Mol Genet Metab. 2020 Nov;131(3):289-298. doi: 10.1016/j.ymgme.2020.10.003. Epub 2020 Oct 10. Mol Genet Metab. 2020. PMID: 33317798 Free PMC article. Review.
-
NAGS, CPS1, and SLC25A13 (Citrin) at the Crossroads of Arginine and Pyrimidines Metabolism in Tumor Cells.Int J Mol Sci. 2023 Apr 4;24(7):6754. doi: 10.3390/ijms24076754. Int J Mol Sci. 2023. PMID: 37047726 Free PMC article.
-
Enhanced hepatic differentiation in the subpopulation of human amniotic stem cells under 3D multicellular microenvironment.World J Stem Cells. 2019 Sep 26;11(9):705-721. doi: 10.4252/wjsc.v11.i9.705. World J Stem Cells. 2019. PMID: 31616545 Free PMC article.
-
Influence of extracellular matrix composition on tumour cell behaviour in a biomimetic in vitro model for hepatocellular carcinoma.Sci Rep. 2023 Jan 13;13(1):748. doi: 10.1038/s41598-023-27997-3. Sci Rep. 2023. PMID: 36639512 Free PMC article.
References
-
- Siddiqui MT, Saboorian MH, Gokaslan ST, et al Diagnostic utility of the HepPar1 antibody to differentiate hepatocellular carcinoma from metastatic carcinoma in fine‐needle aspiration samples. Cancer. 2002; 96: 49–52. - PubMed
-
- Diez‐Fernandez C, Gallego J, Haberle J, et al The study of Carbamoyl Phosphate Synthetase 1 deficiency sheds light on the mechanism for switching on/off the urea cycle. J Genet Genomics. 2015; 42: 249–60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

