Identifying Candidate Genes that Underlie Cellular pH Sensitivity in Serotonin Neurons Using Transcriptomics: A Potential Role for Kir5.1 Channels
- PMID: 28270749
- PMCID: PMC5318415
- DOI: 10.3389/fncel.2017.00034
Identifying Candidate Genes that Underlie Cellular pH Sensitivity in Serotonin Neurons Using Transcriptomics: A Potential Role for Kir5.1 Channels
Abstract
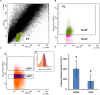
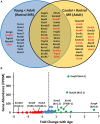
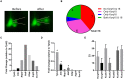
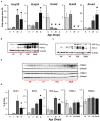
Ventilation is continuously adjusted by a neural network to maintain blood gases and pH. Acute CO2 and/or pH regulation requires neural feedback from brainstem cells that encode CO2/pH to modulate ventilation, including but not limited to brainstem serotonin (5-HT) neurons. Brainstem 5-HT neurons modulate ventilation and are stimulated by hypercapnic acidosis, the sensitivity of which increases with increasing postnatal age. The proper function of brainstem 5-HT neurons, particularly during post-natal development is critical given that multiple abnormalities in the 5-HT system have been identified in victims of Sudden Infant Death Syndrome. Here, we tested the hypothesis that there are age-dependent increases in expression of pH-sensitive ion channels in brainstem 5-HT neurons, which may underlie their cellular CO2/pH sensitivity. Midline raphe neurons were acutely dissociated from neonatal and mature transgenic SSePet-eGFP rats [which have enhanced green fluorescent protein (eGFP) expression in all 5-HT neurons] and sorted with fluorescence-activated cell sorting (FACS) into 5-HT-enriched and non-5-HT cell pools for subsequent RNA extraction, cDNA library preparation and RNA sequencing. Overlapping differential expression analyses pointed to age-dependent shifts in multiple ion channels, including but not limited to the pH-sensitive potassium ion (K+) channel genes kcnj10 (Kir4.1), kcnj16 (Kir5.1), kcnk1 (TWIK-1), kcnk3 (TASK-1) and kcnk9 (TASK-3). Intracellular contents isolated from single adult eGFP+ 5-HT neurons confirmed gene expression of Kir4.1, Kir5.1 and other K+ channels, but also showed heterogeneity in the expression of multiple genes. 5-HT neuron-enriched cell pools from selected post-natal ages showed increases in Kir4.1, Kir5.1, and TWIK-1, fitting with age-dependent increases in Kir4.1 and Kir5.1 protein expression in raphe tissue samples. Immunofluorescence imaging confirmed Kir5.1 protein was co-localized to brainstem neurons and glia including 5-HT neurons as expected. However, Kir4.1 protein expression was restricted to glia, suggesting that it may not contribute to 5-HT neuron pH sensitivity. Although there are caveats to this approach, the data suggest that pH-sensitive Kir5.1 channels may underlie cellular CO2/pH chemosensitivity in brainstem 5-HT neurons.
Keywords: RNA sequencing; chemoreception; control of breathing; potassium channels; serotonin.
Figures

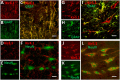
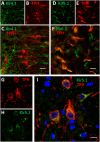
Similar articles
-
Modulation of kir4.1 and kir5.1 by hypercapnia and intracellular acidosis.J Physiol. 2000 May 1;524 Pt 3(Pt 3):725-35. doi: 10.1111/j.1469-7793.2000.00725.x. J Physiol. 2000. PMID: 10790154 Free PMC article.
-
Modulation of the heteromeric Kir4.1-Kir5.1 channel by multiple neurotransmitters via Galphaq-coupled receptors.J Cell Physiol. 2008 Jan;214(1):84-95. doi: 10.1002/jcp.21169. J Cell Physiol. 2008. PMID: 17559083 Free PMC article.
-
Expression and coexpression of CO2-sensitive Kir channels in brainstem neurons of rats.J Membr Biol. 2004 Feb 1;197(3):179-91. doi: 10.1007/s00232-004-0652-4. J Membr Biol. 2004. PMID: 15042349
-
Diverse functions of the inward-rectifying potassium channel Kir5.1 and its relationship with human diseases.Front Physiol. 2023 Feb 27;14:1127893. doi: 10.3389/fphys.2023.1127893. eCollection 2023. Front Physiol. 2023. PMID: 36923292 Free PMC article. Review.
-
Role of inwardly rectifying K+ channel 5.1 (Kir5.1) in the regulation of renal membrane transport.Curr Opin Nephrol Hypertens. 2022 Sep 1;31(5):479-485. doi: 10.1097/MNH.0000000000000817. Epub 2022 Jul 11. Curr Opin Nephrol Hypertens. 2022. PMID: 35894283 Review.
Cited by
-
Perspectives on the basis of seizure-induced respiratory dysfunction.Front Neural Circuits. 2022 Dec 20;16:1033756. doi: 10.3389/fncir.2022.1033756. eCollection 2022. Front Neural Circuits. 2022. PMID: 36605420 Free PMC article. Review.
-
Kir5.1 channels: potential role in epilepsy and seizure disorders.Am J Physiol Cell Physiol. 2022 Sep 1;323(3):C706-C717. doi: 10.1152/ajpcell.00235.2022. Epub 2022 Jul 18. Am J Physiol Cell Physiol. 2022. PMID: 35848616 Free PMC article. Review.
-
Inwardly rectifying potassium channel 5.1: Structure, function, and possible roles in diseases.Genes Dis. 2020 Mar 21;8(3):272-278. doi: 10.1016/j.gendis.2020.03.006. eCollection 2021 May. Genes Dis. 2020. PMID: 33997174 Free PMC article. Review.
-
Kir4.1 Dysfunction in the Pathophysiology of Depression: A Systematic Review.Cells. 2021 Oct 1;10(10):2628. doi: 10.3390/cells10102628. Cells. 2021. PMID: 34685608 Free PMC article.
-
Sex Differences in Biophysical Signatures across Molecularly Defined Medial Amygdala Neuronal Subpopulations.eNeuro. 2020 Jul 2;7(4):ENEURO.0035-20.2020. doi: 10.1523/ENEURO.0035-20.2020. Print 2020 Jul/Aug. eNeuro. 2020. PMID: 32493755 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

