Dynamic proteomics reveals bimodal protein dynamics of cancer cells in response to HSP90 inhibitor
- PMID: 28270142
- PMCID: PMC5341406
- DOI: 10.1186/s12918-017-0410-8
Dynamic proteomics reveals bimodal protein dynamics of cancer cells in response to HSP90 inhibitor
Abstract
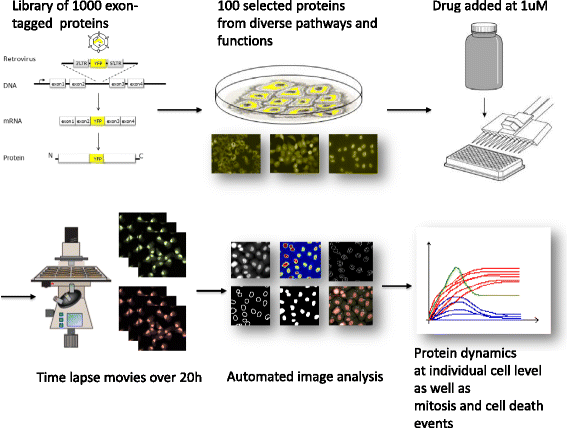
Background: Drugs often kill some cancer cells while others survive. This stochastic outcome is seen even in clonal cells grown under the same conditions. Understanding the molecular reasons for this stochastic outcome is a current challenge, which requires studying the proteome at the single cell level over time. In a previous study we used dynamic proteomics to study the response of cancer cells to a DNA damaging drug, camptothecin. Several proteins showed bimodal dynamics: they rose in some cells and decreased in others, in a way that correlated with eventual cell fate: death or survival. Here we ask whether bimodality is a special case for camptothecin, or whether it occurs for other drugs as well. To address this, we tested a second drug with a different mechanism of action, an HSP90 inhibitor. We used dynamic proteomics to follow 100 proteins in space and time, endogenously tagged in their native chromosomal location in individual living human lung-cancer cells, following drug administration.
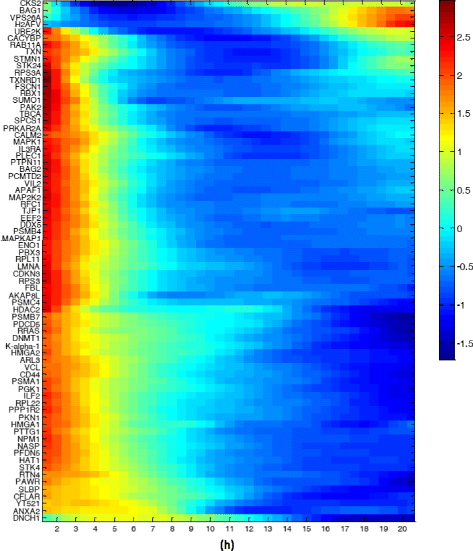
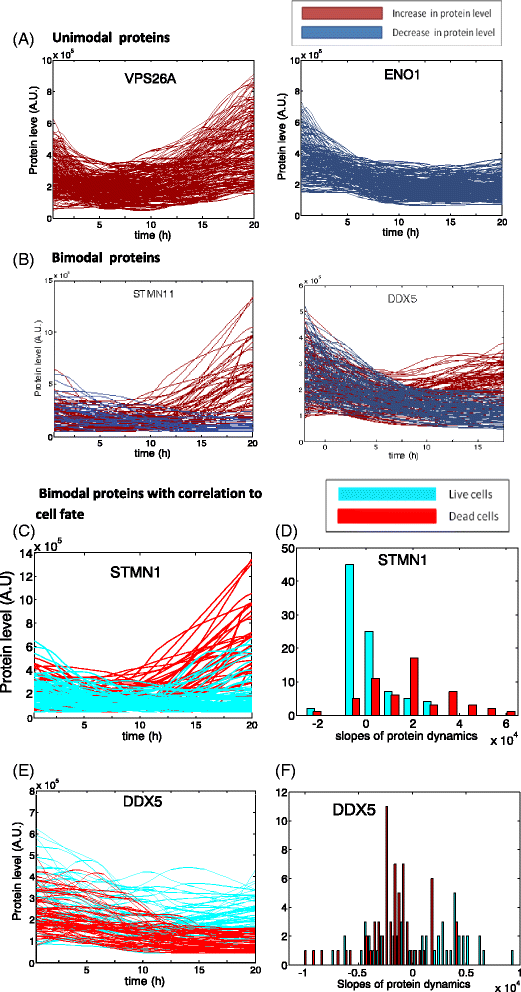
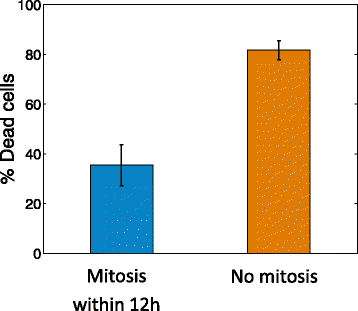
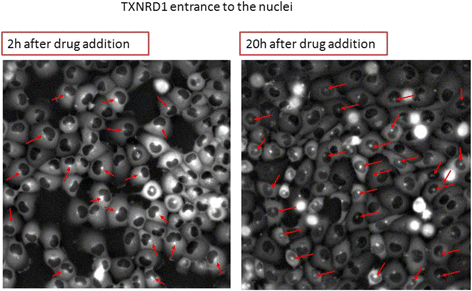
Results: We find bimodal dynamics for a quarter of the proteins. In some cells these proteins strongly rise in level about 12 h after treatment, but in other cells their level drops or remains constant. The proteins which rise in surviving cells included anti-apoptotic factors such as DDX5, and cell cycle regulators such as RFC1. The proteins that rise in cells that eventually die include pro-apoptotic factors such as APAF1. The two drugs shared some aspects in their single-cell response, including 7 of the bimodal proteins and translocation of oxidative response proteins to the nucleus, but differed in other aspects, with HSP90i showing more bimodal proteins. Moreover, the cell cycle phase at drug administration impacted the probability to die from HSP90i but not camptothecin.
Conclusions: Single-cell dynamic proteomics reveals sub-populations of cells within a clonal cell line with different protein dynamics in response to a drug. These different dynamics correlate with cell survival or death. Bimodal proteins which correlate with cell fate may be potential drug targets to enhance the effects of therapy.
Figures
Similar articles
-
Dynamic proteomics of individual cancer cells in response to a drug.Science. 2008 Dec 5;322(5907):1511-6. doi: 10.1126/science.1160165. Epub 2008 Nov 20. Science. 2008. PMID: 19023046
-
Efficacy of Hsp90 inhibition for induction of apoptosis and inhibition of growth in cervical carcinoma cells in vitro and in vivo.Cancer Chemother Pharmacol. 2008 Apr;61(4):669-81. doi: 10.1007/s00280-007-0522-8. Epub 2007 Jun 20. Cancer Chemother Pharmacol. 2008. PMID: 17579866
-
Dynamic impacts of the inhibition of the molecular chaperone Hsp90 on the T-cell proteome have implications for anti-cancer therapy.PLoS One. 2013 Nov 27;8(11):e80425. doi: 10.1371/journal.pone.0080425. eCollection 2013. PLoS One. 2013. PMID: 24312219 Free PMC article.
-
Heat-shock protein 90 (Hsp90) as anticancer target for drug discovery: an ample computational perspective.Chem Biol Drug Des. 2015 Nov;86(5):1131-60. doi: 10.1111/cbdd.12582. Epub 2015 Jun 11. Chem Biol Drug Des. 2015. PMID: 25958815 Review.
-
Targeting mitosis for anti-cancer therapy.BioDrugs. 2007;21(4):225-33. doi: 10.2165/00063030-200721040-00003. BioDrugs. 2007. PMID: 17628120 Review.
Cited by
-
Mechanisms and clinical implications of tumor heterogeneity and convergence on recurrent phenotypes.J Mol Med (Berl). 2017 Nov;95(11):1167-1178. doi: 10.1007/s00109-017-1587-4. Epub 2017 Sep 4. J Mol Med (Berl). 2017. PMID: 28871446 Free PMC article. Review.
-
Heat shock protein 90 promotes RNA helicase DDX5 accumulation and exacerbates hepatocellular carcinoma by inhibiting autophagy.Cancer Biol Med. 2021 Mar 25;18(3):693-704. doi: 10.20892/j.issn.2095-3941.2020.0262. Online ahead of print. Cancer Biol Med. 2021. PMID: 33764710 Free PMC article.
-
Synthesis and preliminary evaluation of octreotate conjugates of bioactive synthetic amatoxins for targeting somatostatin receptor (sstr2) expressing cells.RSC Chem Biol. 2021 Oct 7;3(1):69-78. doi: 10.1039/d1cb00036e. eCollection 2022 Jan 5. RSC Chem Biol. 2021. PMID: 35128410 Free PMC article.
-
Human Protein-l-isoaspartate O-Methyltransferase Domain-Containing Protein 1 (PCMTD1) Associates with Cullin-RING Ligase Proteins.Biochemistry. 2022 May 17;61(10):879-894. doi: 10.1021/acs.biochem.2c00130. Epub 2022 Apr 29. Biochemistry. 2022. PMID: 35486881 Free PMC article.
-
Identification of Predictive Biomarkers of Response to HSP90 Inhibitors in Lung Adenocarcinoma.Int J Mol Sci. 2021 Mar 3;22(5):2538. doi: 10.3390/ijms22052538. Int J Mol Sci. 2021. PMID: 33802597 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

