Machine Learning of Global Phosphoproteomic Profiles Enables Discrimination of Direct versus Indirect Kinase Substrates
- PMID: 28265048
- PMCID: PMC5417821
- DOI: 10.1074/mcp.M116.066233
Machine Learning of Global Phosphoproteomic Profiles Enables Discrimination of Direct versus Indirect Kinase Substrates
Abstract
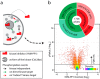
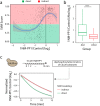
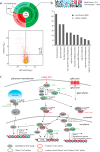
Mass spectrometry allows quantification of tens of thousands of phosphorylation sites from minute amounts of cellular material. Despite this wealth of information, our understanding of phosphorylation-based signaling is limited, in part because it is not possible to deconvolute substrate phosphorylation that is directly mediated by a particular kinase versus phosphorylation that is mediated by downstream kinases. Here, we describe a framework for assignment of direct in vivo kinase substrates using a combination of selective chemical inhibition, quantitative phosphoproteomics, and machine learning techniques. Our workflow allows classification of phosphorylation events following inhibition of an analog-sensitive kinase into kinase-independent effects of the inhibitor, direct effects on cognate substrates, and indirect effects mediated by downstream kinases or phosphatases. We applied this method to identify many direct targets of Cdc28 and Snf1 kinases in the budding yeast Saccharomyces cerevisiae Global phosphoproteome analysis of acute time-series demonstrated that dephosphorylation of direct kinase substrates occurs more rapidly compared with indirect substrates, both after inhibitor treatment and under a physiological nutrient shift in wt cells. Mutagenesis experiments revealed a high proportion of functionally relevant phosphorylation sites on Snf1 targets. For example, Snf1 itself was inhibited through autophosphorylation on Ser391 and new phosphosites were discovered that modulate the activity of the Reg1 regulatory subunit of the Glc7 phosphatase and the Gal83 β-subunit of SNF1 complex. This methodology applies to any kinase for which a functional analog sensitive version can be constructed to facilitate the dissection of the global phosphorylation network.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
Reg1 protein regulates phosphorylation of all three Snf1 isoforms but preferentially associates with the Gal83 isoform.Eukaryot Cell. 2011 Dec;10(12):1628-36. doi: 10.1128/EC.05176-11. Epub 2011 Oct 14. Eukaryot Cell. 2011. PMID: 22002657 Free PMC article.
-
PP1 phosphatase-binding motif in Reg1 protein of Saccharomyces cerevisiae is required for interaction with both the PP1 phosphatase Glc7 and the Snf1 protein kinase.Cell Signal. 2010 Jul;22(7):1013-21. doi: 10.1016/j.cellsig.2010.02.003. Epub 2010 Feb 17. Cell Signal. 2010. PMID: 20170726 Free PMC article.
-
Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase.J Biol Chem. 2008 Jan 4;283(1):222-230. doi: 10.1074/jbc.M707957200. Epub 2007 Nov 8. J Biol Chem. 2008. PMID: 17991748 Free PMC article.
-
Illuminating the dark phosphoproteome.Sci Signal. 2019 Jan 22;12(565):eaau8645. doi: 10.1126/scisignal.aau8645. Sci Signal. 2019. PMID: 30670635 Review.
-
Phosphoproteomic Approaches for Identifying Phosphatase and Kinase Substrates.Molecules. 2023 Apr 24;28(9):3675. doi: 10.3390/molecules28093675. Molecules. 2023. PMID: 37175085 Free PMC article. Review.
Cited by
-
The Mycobacterium tuberculosis protein O-phosphorylation landscape.Nat Microbiol. 2023 Mar;8(3):548-561. doi: 10.1038/s41564-022-01313-7. Epub 2023 Jan 23. Nat Microbiol. 2023. PMID: 36690861 Free PMC article.
-
Delineating the contribution of Spc105-bound PP1 to spindle checkpoint silencing and kinetochore microtubule attachment regulation.J Cell Biol. 2019 Dec 2;218(12):3926-3942. doi: 10.1083/jcb.201810172. Epub 2019 Oct 24. J Cell Biol. 2019. PMID: 31649151 Free PMC article.
-
Pharmacological approaches to understanding protein kinase signaling networks.Front Pharmacol. 2023 Dec 14;14:1310135. doi: 10.3389/fphar.2023.1310135. eCollection 2023. Front Pharmacol. 2023. PMID: 38164473 Free PMC article. Review.
-
Snf1 cooperates with the CWI MAPK pathway to mediate the degradation of Med13 following oxidative stress.Microb Cell. 2018 Jun 25;5(8):357-370. doi: 10.15698/mic2018.08.641. Microb Cell. 2018. PMID: 30175106 Free PMC article.
-
Synthesizing Signaling Pathways from Temporal Phosphoproteomic Data.Cell Rep. 2018 Sep 25;24(13):3607-3618. doi: 10.1016/j.celrep.2018.08.085. Cell Rep. 2018. PMID: 30257219 Free PMC article.
References
-
- Pawson T., and Scott J. D. (2005) Protein phosphorylation in signaling—50 years and counting. Trends Biochem. Sci. 30, 286–290 - PubMed
-
- Hunter T. (2000) Signaling—2000 and beyond. Cell 100, 113–127 - PubMed
-
- Hunter T., and Plowman G. D. (1997) The protein kinases of budding yeast: Six score and more. Trends Biochem. Sci. 22, 18–22 - PubMed
-
- Macek B., Mann M., and Olsen J. V. (2009) Global and site-specific quantitative phosphoproteomics: Principles and applications. Annu. Rev. Pharmacol. Toxicol. 49, 199–221 - PubMed
-
- Rigbolt K. T., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., and Blagoev B. (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4, rs3. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

