The dev Operon Regulates the Timing of Sporulation during Myxococcus xanthus Development
- PMID: 28264995
- PMCID: PMC5405206
- DOI: 10.1128/JB.00788-16
The dev Operon Regulates the Timing of Sporulation during Myxococcus xanthus Development
Abstract
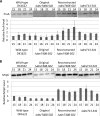
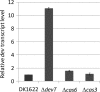
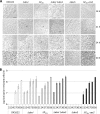
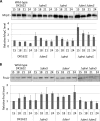
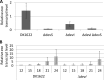
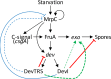
Myxococcus xanthus undergoes multicellular development when starved. Thousands of rod-shaped cells coordinate their movements and aggregate into mounds in which cells differentiate into spores. Mutations in the dev operon impair development. The dev operon encompasses a clustered regularly interspaced short palindromic repeat-associated (CRISPR-Cas) system. Null mutations in devI, a small gene at the beginning of the dev operon, suppress the developmental defects caused by null mutations in the downstream devR and devS genes but failed to suppress defects caused by a small in-frame deletion in devT We provide evidence that the original mutant has a second-site mutation. We show that devT null mutants exhibit developmental defects indistinguishable from devR and devS null mutants, and a null mutation in devI suppresses the defects of a devT null mutation. The similarity of DevTRS proteins to components of the CRISPR-associated complex for antiviral defense (Cascade), together with our molecular characterization of dev mutants, support a model in which DevTRS form a Cascade-like subcomplex that negatively autoregulates dev transcript accumulation and prevents DevI overproduction that would strongly inhibit sporulation. Our results also suggest that DevI transiently inhibits sporulation when regulated normally. The mechanism of transient inhibition may involve MrpC, a key transcription factor, whose translation appears to be weakly inhibited by DevI. Finally, our characterization of a devI devS mutant indicates that very little exo transcript is required for sporulation, which is surprising since Exo proteins help form the polysaccharide spore coat.IMPORTANCE CRISPR-Cas systems typically function as adaptive immune systems in bacteria. The dev CRISPR-Cas system of M. xanthus has been proposed to prevent bacteriophage infection during development, but how dev controls sporulation has been elusive. Recent evidence supported a model in which DevR and DevS prevent overproduction of DevI, a predicted 40-residue inhibitor of sporulation. We provide genetic evidence that DevT functions together with DevR and DevS to prevent DevI overproduction. We also show that spores form about 6 h earlier in mutants lacking devI than in the wild type. Only a minority of natural isolates appear to have a functional dev promoter and devI, suggesting that a functional dev CRISPR-Cas system evolved recently in niches where delayed sporulation and/or protection from bacteriophage infection proved advantageous.
Keywords: CRISPR-Cas; Myxococcus xanthus; bacterial development; dev operon; gene regulation; signaling; sporulation.
Copyright © 2017 American Society for Microbiology.
Figures


Similar articles
-
devI is an evolutionarily young negative regulator of Myxococcus xanthus development.J Bacteriol. 2015 Apr;197(7):1249-62. doi: 10.1128/JB.02542-14. Epub 2015 Feb 2. J Bacteriol. 2015. PMID: 25645563 Free PMC article.
-
Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats.J Bacteriol. 2007 May;189(10):3738-50. doi: 10.1128/JB.00187-07. Epub 2007 Mar 16. J Bacteriol. 2007. PMID: 17369305 Free PMC article.
-
Regulation of late-acting operons by three transcription factors and a CRISPR-Cas component during Myxococcus xanthus development.Mol Microbiol. 2024 May;121(5):1002-1020. doi: 10.1111/mmi.15252. Epub 2024 Mar 25. Mol Microbiol. 2024. PMID: 38525557
-
Highly Signal-Responsive Gene Regulatory Network Governing Myxococcus Development.Trends Genet. 2017 Jan;33(1):3-15. doi: 10.1016/j.tig.2016.10.006. Epub 2016 Dec 2. Trends Genet. 2017. PMID: 27916428 Free PMC article. Review.
-
Dual regulation with Ser/Thr kinase cascade and a His/Asp TCS in Myxococcus xanthus.Adv Exp Med Biol. 2008;631:111-21. doi: 10.1007/978-0-387-78885-2_7. Adv Exp Med Biol. 2008. PMID: 18792684 Review.
Cited by
-
Bacteriophages of Myxococcus xanthus, a Social Bacterium.Viruses. 2018 Jul 18;10(7):374. doi: 10.3390/v10070374. Viruses. 2018. PMID: 30021959 Free PMC article. Review.
-
A Simple Criterion for Inferring CRISPR Array Direction.Front Microbiol. 2019 Sep 4;10:2054. doi: 10.3389/fmicb.2019.02054. eCollection 2019. Front Microbiol. 2019. PMID: 31551987 Free PMC article.
-
Multifactorial control of the expression of a CRISPR-Cas system by an extracytoplasmic function σ/anti-σ pair and a global regulatory complex.Nucleic Acids Res. 2018 Jul 27;46(13):6726-6745. doi: 10.1093/nar/gky475. Nucleic Acids Res. 2018. PMID: 29893914 Free PMC article.
-
Alternative functions of CRISPR-Cas systems in the evolutionary arms race.Nat Rev Microbiol. 2022 Jun;20(6):351-364. doi: 10.1038/s41579-021-00663-z. Epub 2022 Jan 6. Nat Rev Microbiol. 2022. PMID: 34992260 Review.
-
Evolutionary plasticity and functional versatility of CRISPR systems.PLoS Biol. 2022 Jan 5;20(1):e3001481. doi: 10.1371/journal.pbio.3001481. eCollection 2022 Jan. PLoS Biol. 2022. PMID: 34986140 Free PMC article.
References
-
- Yang Z, Higgs P. 2014. Myxobacteria: genomics, cellular and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
-
- Lee B, Holkenbrink C, Treuner-Lange A, Higgs PI. 2012. Myxococcus xanthus developmental cell fate production: heterogeneous accumulation of developmental regulatory proteins and reexamination of the role of MazF in developmental lysis. J Bacteriol 194:3058–3068. doi:10.1128/JB.06756-11. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

