Importance of ROS-mediated autophagy in determining apoptotic cell death induced by physapubescin B
- PMID: 28258023
- PMCID: PMC5333534
- DOI: 10.1016/j.redox.2017.02.017
Importance of ROS-mediated autophagy in determining apoptotic cell death induced by physapubescin B
Abstract
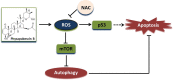
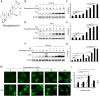
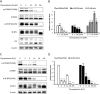
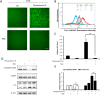
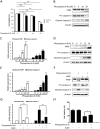
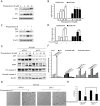
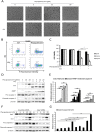
Physapubescin B, a steroidal compound extracted from the plant Physalis pubescens L. (Solanaceae), has been reported to possess anti-cancer potential, whereas the molecular mechanism remains elusive. In this study, we first demonstrated that physapubescin B induced autophagy in human cancer cells based on the evidence that physapubescin B increased lipidation of microtubule-associated protein 1 light chain 3 (LC3) as well as number of GFP-LC3 puncta. We further examined the molecular mechanisms and found that physapubescin B enhanced the autophagic flux through promotion of reactive oxygen species (ROS)-mediated suppression of mammalian target of rapamycin complex I (mTORC1), the key negative regulator of autophagy. Additionally, excessive ROS caused by physapubescin B also induced p53-dependent apoptotic cell death. Furthermore, we provided evidence that inhibition of autophagy either by a chemical inhibitor or gene silencing promoted physapubescin B-induced apoptotic cell death, indicating that autophagy serves as a cell survival mechanism to protect cell death. Thus, our data provide a clue that inhibition of autophagy would serve as a novel strategy for enhancing the anti-cancer potential of physapubescin B.
Keywords: Apoptotic cell death; Autophagy; MTORC1; Physapubescin B; ROS.
Copyright © 2017. Published by Elsevier B.V.
Figures
Similar articles
-
Physalin A induces apoptosis via p53-Noxa-mediated ROS generation, and autophagy plays a protective role against apoptosis through p38-NF-κB survival pathway in A375-S2 cells.J Ethnopharmacol. 2013 Jul 9;148(2):544-55. doi: 10.1016/j.jep.2013.04.051. Epub 2013 May 14. J Ethnopharmacol. 2013. PMID: 23684722
-
Physapubescin B Exhibits Potent Activity against Human Prostate Cancer In Vitro and In Vivo.J Agric Food Chem. 2015 Nov 4;63(43):9504-12. doi: 10.1021/acs.jafc.5b03045. Epub 2015 Oct 21. J Agric Food Chem. 2015. PMID: 26415552
-
Walsuronoid B induces mitochondrial and lysosomal dysfunction leading to apoptotic rather than autophagic cell death via ROS/p53 signaling pathways in liver cancer.Biochem Pharmacol. 2017 Oct 15;142:71-86. doi: 10.1016/j.bcp.2017.06.134. Epub 2017 Jul 1. Biochem Pharmacol. 2017. PMID: 28673807
-
Chalcone flavokawain B induces autophagic-cell death via reactive oxygen species-mediated signaling pathways in human gastric carcinoma and suppresses tumor growth in nude mice.Arch Toxicol. 2017 Oct;91(10):3341-3364. doi: 10.1007/s00204-017-1967-0. Epub 2017 Apr 3. Arch Toxicol. 2017. PMID: 28374157
-
BIX-01294 induces autophagy-associated cell death via EHMT2/G9a dysfunction and intracellular reactive oxygen species production.Autophagy. 2013 Dec;9(12):2126-39. doi: 10.4161/auto.26308. Autophagy. 2013. PMID: 24322755
Cited by
-
Autophagy-dependent apoptosis is triggered by a semi-synthetic [6]-gingerol analogue in triple negative breast cancer cells.Oncotarget. 2018 Jul 20;9(56):30787-30804. doi: 10.18632/oncotarget.25704. eCollection 2018 Jul 20. Oncotarget. 2018. PMID: 30112107 Free PMC article.
-
Calpain 6 inhibits autophagy in inflammatory environments: A preliminary study on myoblasts and a chronic kidney disease rat model.Int J Mol Med. 2021 Oct;48(4):194. doi: 10.3892/ijmm.2021.5027. Epub 2021 Aug 26. Int J Mol Med. 2021. PMID: 34435644 Free PMC article.
-
The Current Status of the Pharmaceutical Potential of Juniperus L. Metabolites.Medicines (Basel). 2018 Jul 31;5(3):81. doi: 10.3390/medicines5030081. Medicines (Basel). 2018. PMID: 30065158 Free PMC article. Review.
-
p53 Modulation of Autophagy Signaling in Cancer Therapies: Perspectives Mechanism and Therapeutic Targets.Front Cell Dev Biol. 2022 Jan 26;10:761080. doi: 10.3389/fcell.2022.761080. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35155422 Free PMC article. Review.
-
Anticancer Mechanisms of Bioactive Compounds from Solanaceae: An Update.Cancers (Basel). 2021 Oct 5;13(19):4989. doi: 10.3390/cancers13194989. Cancers (Basel). 2021. PMID: 34638473 Free PMC article. Review.
References
-
- Huang Y., Cui J., Chen S., Gan C., Zhou A. Synthesis and antiproliferative activity of some steroidal lactams. Steroids. 2011;76:1346–1350. - PubMed
-
- Ji L., Yuan Y., Luo L., Chen Z., Ma X. Physalins with anti-inflammatory activity are present in Physalis alkekengi var. franchetii and can function as Michael reaction acceptors. Steroids. 2012;77:441–447. - PubMed
-
- Yang B.-Y., Guo R., Li T., Wu J.-J., Zhang J. New anti-inflammatory withanolides from the leaves of Datura metel L. Steroids. 2014;87:26–34. - PubMed
-
- Silva M.T., Simas S.M., Batista T.G., Cardarelli P., Tomassini T.C. Studies on antimicrobial activity, in vitro, of Physalis angulata L.(Solanaceae) fraction and physalin B bringing out the importance of assay determination. Mem. Inst. Oswaldo Cruz. 2005;100(7):779–782. - PubMed
-
- Yang Y.K., Xie S.D., Xu W.X., Nian Y., Liu X.L. Six new physalins from Physalis alkekengi var. franchetii and their cytotoxicity and antibacterial activity. Fitoterapia. 2016;112:144–152. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

