Integrated systems biology analysis of KSHV latent infection reveals viral induction and reliance on peroxisome mediated lipid metabolism
- PMID: 28257516
- PMCID: PMC5352148
- DOI: 10.1371/journal.ppat.1006256
Integrated systems biology analysis of KSHV latent infection reveals viral induction and reliance on peroxisome mediated lipid metabolism
Abstract
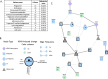
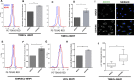
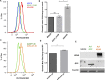
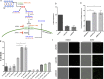
Kaposi's Sarcoma associated Herpesvirus (KSHV), an oncogenic, human gamma-herpesvirus, is the etiological agent of Kaposi's Sarcoma the most common tumor of AIDS patients world-wide. KSHV is predominantly latent in the main KS tumor cell, the spindle cell, a cell of endothelial origin. KSHV modulates numerous host cell-signaling pathways to activate endothelial cells including major metabolic pathways involved in lipid metabolism. To identify the underlying cellular mechanisms of KSHV alteration of host signaling and endothelial cell activation, we identified changes in the host proteome, phosphoproteome and transcriptome landscape following KSHV infection of endothelial cells. A Steiner forest algorithm was used to integrate the global data sets and, together with transcriptome based predicted transcription factor activity, cellular networks altered by latent KSHV were predicted. Several interesting pathways were identified, including peroxisome biogenesis. To validate the predictions, we showed that KSHV latent infection increases the number of peroxisomes per cell. Additionally, proteins involved in peroxisomal lipid metabolism of very long chain fatty acids, including ABCD3 and ACOX1, are required for the survival of latently infected cells. In summary, novel cellular pathways altered during herpesvirus latency that could not be predicted by a single systems biology platform, were identified by integrated proteomics and transcriptomics data analysis and when correlated with our metabolomics data revealed that peroxisome lipid metabolism is essential for KSHV latent infection of endothelial cells.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures



Similar articles
-
Glycolysis, Glutaminolysis, and Fatty Acid Synthesis Are Required for Distinct Stages of Kaposi's Sarcoma-Associated Herpesvirus Lytic Replication.J Virol. 2017 Apr 28;91(10):e02237-16. doi: 10.1128/JVI.02237-16. Print 2017 May 15. J Virol. 2017. PMID: 28275189 Free PMC article.
-
Activation of cellular metabolism during latent Kaposi's Sarcoma herpesvirus infection.Curr Opin Virol. 2016 Aug;19:45-9. doi: 10.1016/j.coviro.2016.06.012. Epub 2016 Jul 18. Curr Opin Virol. 2016. PMID: 27434732 Free PMC article. Review.
-
Latent KSHV infection of endothelial cells induces integrin beta3 to activate angiogenic phenotypes.PLoS Pathog. 2011 Dec;7(12):e1002424. doi: 10.1371/journal.ppat.1002424. Epub 2011 Dec 8. PLoS Pathog. 2011. PMID: 22174684 Free PMC article.
-
STAT3 Regulates Lytic Activation of Kaposi's Sarcoma-Associated Herpesvirus.J Virol. 2015 Nov;89(22):11347-55. doi: 10.1128/JVI.02008-15. Epub 2015 Sep 2. J Virol. 2015. PMID: 26339061 Free PMC article.
-
Kaposi's sarcoma herpesvirus-induced endothelial cell reprogramming supports viral persistence and contributes to Kaposi's sarcoma tumorigenesis.Curr Opin Virol. 2017 Oct;26:156-162. doi: 10.1016/j.coviro.2017.09.002. Epub 2017 Oct 12. Curr Opin Virol. 2017. PMID: 29031103 Review.
Cited by
-
Peroxisomes in Immune Response and Inflammation.Int J Mol Sci. 2019 Aug 8;20(16):3877. doi: 10.3390/ijms20163877. Int J Mol Sci. 2019. PMID: 31398943 Free PMC article. Review.
-
Peroxisomes support human herpesvirus 8 latency by stabilizing the viral oncogenic protein vFLIP via the MAVS-TRAF complex.PLoS Pathog. 2018 May 10;14(5):e1007058. doi: 10.1371/journal.ppat.1007058. eCollection 2018 May. PLoS Pathog. 2018. PMID: 29746593 Free PMC article.
-
Hepatitis C virus alters the morphology and function of peroxisomes.Front Microbiol. 2023 Sep 21;14:1254728. doi: 10.3389/fmicb.2023.1254728. eCollection 2023. Front Microbiol. 2023. PMID: 37808318 Free PMC article.
-
Re-Examining Rotavirus Innate Immune Evasion: Potential Applications of the Reverse Genetics System.mBio. 2022 Aug 30;13(4):e0130822. doi: 10.1128/mbio.01308-22. Epub 2022 Jun 14. mBio. 2022. PMID: 35699371 Free PMC article. Review.
-
Bone Marrow-Derived SH-SY5Y Neuroblastoma Cells Infected with Kaposi's Sarcoma-Associated Herpesvirus Display Unique Infection Phenotypes and Growth Properties.J Virol. 2021 Jun 10;95(13):e0000321. doi: 10.1128/JVI.00003-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33853962 Free PMC article.
References
-
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266(5192):1865–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

