Conformational States of a Soluble, Uncleaved HIV-1 Envelope Trimer
- PMID: 28250125
- PMCID: PMC5411591
- DOI: 10.1128/JVI.00175-17
Conformational States of a Soluble, Uncleaved HIV-1 Envelope Trimer
Abstract
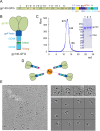
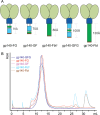
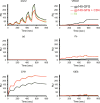
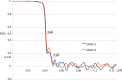
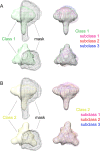
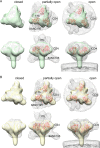
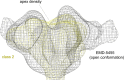
The HIV-1 envelope spike [Env; trimeric (gp160)3 cleaved to (gp120/gp41)3] induces membrane fusion, leading to viral entry. It is also the viral component targeted by neutralizing antibodies. Vaccine development requires production, in quantities suitable for clinical studies, of a recombinant form that resembles functional Env. HIV-1 gp140 trimers-the uncleaved ectodomains of (gp160)3-from a few selected viral isolates adopt a compact conformation with many antigenic properties of native Env spikes. One is currently being evaluated in a clinical trial. We report here low-resolution (20 Å) electron cryomicroscopy (cryoEM) structures of this gp140 trimer, which adopts two principal conformations, one closed and the other slightly open. The former is indistinguishable at this resolution from those adopted by a stabilized, cleaved trimer (SOSIP) or by a membrane-bound Env trimer with a truncated cytoplasmic tail (EnvΔCT). The latter conformation is closer to a partially open Env trimer than to the fully open conformation induced by CD4. These results show that a stable, uncleaved HIV-1 gp140 trimer has a compact structure close to that of native Env.IMPORTANCE Development of any HIV vaccine with a protein component (for either priming or boosting) requires production of a recombinant form to mimic the trimeric, functional HIV-1 envelope spike in quantities suitable for clinical studies. Our understanding of the envelope structure has depended in part on a cleaved, soluble trimer, known as SOSIP.664, stabilized by several modifications, including an engineered disulfide. This construct, which is difficult to produce in large quantities, has yet to induce better antibody responses than those to other envelope-based immunogens, even in animal models. The uncleaved ectodomain of the envelope protein, called gp140, has also been made as a soluble form to mimic the native Env present on the virion surface. Most HIV-1 gp140 preparations are not stable, however, and have an inhomogeneous conformation. The results presented here show that gp140 preparations from suitable isolates can adopt a compact, native-like structure, supporting its use as a vaccine candidate.
Keywords: cryoEM; envelope; human immunodeficiency virus; immunogen.
Copyright © 2017 American Society for Microbiology.
Figures



Similar articles
-
Comparison of Uncleaved and Mature Human Immunodeficiency Virus Membrane Envelope Glycoprotein Trimers.J Virol. 2018 May 29;92(12):e00277-18. doi: 10.1128/JVI.00277-18. Print 2018 Jun 15. J Virol. 2018. PMID: 29618643 Free PMC article.
-
A New Approach to Produce HIV-1 Envelope Trimers: BOTH CLEAVAGE AND PROPER GLYCOSYLATION ARE ESSENTIAL TO GENERATE AUTHENTIC TRIMERS.J Biol Chem. 2015 Aug 7;290(32):19780-95. doi: 10.1074/jbc.M115.656611. Epub 2015 Jun 18. J Biol Chem. 2015. PMID: 26088135 Free PMC article.
-
Stable, uncleaved HIV-1 envelope glycoprotein gp140 forms a tightly folded trimer with a native-like structure.Proc Natl Acad Sci U S A. 2014 Dec 30;111(52):18542-7. doi: 10.1073/pnas.1422269112. Epub 2014 Dec 15. Proc Natl Acad Sci U S A. 2014. PMID: 25512514 Free PMC article.
-
HIV-1 Envelope Conformation, Allostery, and Dynamics.Viruses. 2021 May 7;13(5):852. doi: 10.3390/v13050852. Viruses. 2021. PMID: 34067073 Free PMC article. Review.
-
Stabilizing HIV-1 envelope glycoprotein trimers to induce neutralizing antibodies.Retrovirology. 2018 Sep 12;15(1):63. doi: 10.1186/s12977-018-0445-y. Retrovirology. 2018. PMID: 30208933 Free PMC article. Review.
Cited by
-
Structure and Immune Recognition of the HIV Glycan Shield.Annu Rev Biophys. 2018 May 20;47:499-523. doi: 10.1146/annurev-biophys-060414-034156. Epub 2018 Mar 29. Annu Rev Biophys. 2018. PMID: 29595997 Free PMC article.
-
Improving the Expression and Purification of Soluble, Recombinant Native-Like HIV-1 Envelope Glycoprotein Trimers by Targeted Sequence Changes.J Virol. 2017 May 26;91(12):e00264-17. doi: 10.1128/JVI.00264-17. Print 2017 Jun 15. J Virol. 2017. PMID: 28381572 Free PMC article.
-
Accelerated Clearance and Degradation of Cell-Free HIV by Neutralizing Antibodies Occurs via FcγRIIb on Liver Sinusoidal Endothelial Cells by Endocytosis.J Immunol. 2021 Mar 15;206(6):1284-1296. doi: 10.4049/jimmunol.2000772. Epub 2021 Feb 10. J Immunol. 2021. PMID: 33568400 Free PMC article.
-
Meeting Review: 2018 International Workshop on Structure and Function of the Lentiviral gp41 Cytoplasmic Tail.Viruses. 2018 Nov 7;10(11):613. doi: 10.3390/v10110613. Viruses. 2018. PMID: 30405009 Free PMC article.
-
Neutralizing Antibody Responses following Long-Term Vaccination with HIV-1 Env gp140 in Guinea Pigs.J Virol. 2018 Jun 13;92(13):e00369-18. doi: 10.1128/JVI.00369-18. Print 2018 Jul 1. J Virol. 2018. PMID: 29643249 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

