Bead Based Proteome Enrichment Enhances Features of the Protein Elution Plate (PEP) for Functional Proteomic Profiling
- PMID: 28248280
- PMCID: PMC5217392
- DOI: 10.3390/proteomes3040454
Bead Based Proteome Enrichment Enhances Features of the Protein Elution Plate (PEP) for Functional Proteomic Profiling
Abstract
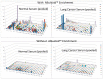
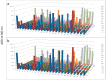
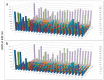
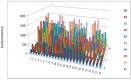
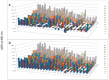
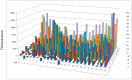
A novel functional proteomics technology called PEP(Protein Elution Plate) was developed to separate complex proteomes from natural sources and analyze protein functions systematically. The technology takes advantage of the powerful resolution of two-dimensional gel electrophoresis (2-D Gels). The modification of electrophoretic conditions in combination with a high-resolution protein elution plate supports the recovery of functionally active proteins. As 2DE(2-Dimensional Electrophoresis) resolution can be limited by protein load, we investigated the use of bead based enrichment technologies, called AlbuVoid™ and KinaSorb™ to determine their effect on the proteomic features which can be generated from the PEP platform. Using a variety of substrates and enzyme activity assays, we report on the benefits of combining bead based enrichment to improve the signal report and the features generated for Hexokinase, Protein Kinase, Protease, and Alkaline Phosphatase activities. As a result, the PEP technology allows systematic analysis of large enzyme families and can build a comprehensive picture of protein function from a complex proteome, providing biological insights that could otherwise not be observed if only protein abundances were analyzed.
Keywords: 2-D gel; PEP; biomarkers; cancer; functional proteomics; protein enrichment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Identification of functional metabolic biomarkers from lung cancer patient serum using PEP technology.Biomark Res. 2016 Jun 1;4:11. doi: 10.1186/s40364-016-0065-4. eCollection 2016. Biomark Res. 2016. PMID: 27252855 Free PMC article.
-
Systematic Glycolytic Enzyme Activity Analysis from Human Serum with PEP Technology.Methods Mol Biol. 2019;1871:69-81. doi: 10.1007/978-1-4939-8814-3_4. Methods Mol Biol. 2019. PMID: 30276732
-
Identification of multiple metabolic enzymes from mice cochleae tissue using a novel functional proteomics technology.PLoS One. 2015 Mar 26;10(3):e0121826. doi: 10.1371/journal.pone.0121826. eCollection 2015. PLoS One. 2015. PMID: 25811366 Free PMC article.
-
Two-dimensional electrophoresis in proteome expression analysis.J Chromatogr B Analyt Technol Biomed Life Sci. 2007 Apr 15;849(1-2):190-202. doi: 10.1016/j.jchromb.2006.11.049. Epub 2006 Dec 22. J Chromatogr B Analyt Technol Biomed Life Sci. 2007. PMID: 17188947 Review.
-
Clinical proteomics in cancer research-promises and limitations of current two-dimensional gel electrophoresis.Curr Med Chem. 2008;15(23):2393-400. doi: 10.2174/092986708785909102. Curr Med Chem. 2008. PMID: 18855668 Review.
Cited by
-
Proteases: Pivot Points in Functional Proteomics.Methods Mol Biol. 2019;1871:313-392. doi: 10.1007/978-1-4939-8814-3_20. Methods Mol Biol. 2019. PMID: 30276748 Free PMC article. Review.
-
Identification of potential serum biomarkers for breast cancer using a functional proteomics technology.Biomark Res. 2017 Mar 14;5:11. doi: 10.1186/s40364-017-0092-9. eCollection 2017. Biomark Res. 2017. PMID: 28293426 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

