Pneumococcal galactose catabolism is controlled by multiple regulators acting on pyruvate formate lyase
- PMID: 28240278
- PMCID: PMC5327383
- DOI: 10.1038/srep43587
Pneumococcal galactose catabolism is controlled by multiple regulators acting on pyruvate formate lyase
Abstract
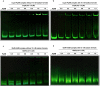
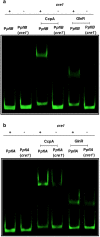
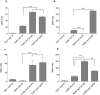
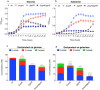
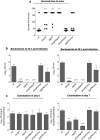
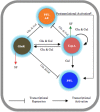
Catabolism of galactose by Streptococcus pneumoniae alters the microbe's metabolism from homolactic to mixed acid fermentation, and this shift is linked to the microbe's virulence. However, the genetic basis of this switch is unknown. Pyruvate formate lyase (PFL) is a crucial enzyme for mixed acid fermentation. Functional PFL requires the activities of two enzymes: pyruvate formate lyase activating enzyme (coded by pflA) and pyruvate formate lyase (coded by pflB). To understand the genetic basis of mixed acid fermentation, transcriptional regulation of pflA and pflB was studied. By microarray analysis of ΔpflB, differential regulation of several transcriptional regulators were identified, and CcpA, and GlnR's role in active PFL synthesis was studied in detail as these regulators directly interact with the putative promoters of both pflA and pflB, their mutation attenuated pneumococcal growth, and their expression was induced on host-derived sugars, indicating that these regulators have a role in sugar metabolism, and multiple regulators are involved in active PFL synthesis. We also found that the influence of each regulator on pflA and pflB expression was distinct in terms of activation and repression, and environmental condition. These results show that active PFL synthesis is finely tuned, and feed-back inhibition and activation are involved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Pyruvate formate lyase is required for pneumococcal fermentative metabolism and virulence.Infect Immun. 2009 Dec;77(12):5418-27. doi: 10.1128/IAI.00178-09. Epub 2009 Sep 14. Infect Immun. 2009. PMID: 19752030 Free PMC article.
-
Molecular characterization of CcpA and involvement of this protein in transcriptional regulation of lactate dehydrogenase and pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis.Appl Environ Microbiol. 2004 Sep;70(9):5244-51. doi: 10.1128/AEM.70.9.5244-5251.2004. Appl Environ Microbiol. 2004. PMID: 15345406 Free PMC article.
-
The Rgg1518 transcriptional regulator is a necessary facet of sugar metabolism and virulence in Streptococcus pneumoniae.Mol Microbiol. 2021 Sep;116(3):996-1008. doi: 10.1111/mmi.14788. Epub 2021 Aug 10. Mol Microbiol. 2021. PMID: 34328238 Free PMC article.
-
A radical-chemical route to acetyl-CoA: the anaerobically induced pyruvate formate-lyase system of Escherichia coli.FEMS Microbiol Rev. 1990 Aug;6(4):383-98. doi: 10.1111/j.1574-6968.1990.tb04108.x. FEMS Microbiol Rev. 1990. PMID: 2248795 Review.
-
A glycyl radical solution: oxygen-dependent interconversion of pyruvate formate-lyase.Mol Microbiol. 1998 Aug;29(4):945-54. doi: 10.1046/j.1365-2958.1998.00941.x. Mol Microbiol. 1998. PMID: 9767563 Review.
Cited by
-
Structure-function analysis for the development of peptide inhibitors for a Gram-positive quorum sensing system.Mol Microbiol. 2022 Jun;117(6):1464-1478. doi: 10.1111/mmi.14921. Epub 2022 May 28. Mol Microbiol. 2022. PMID: 35575437 Free PMC article.
-
TprA/PhrA Quorum Sensing System Has a Major Effect on Pneumococcal Survival in Respiratory Tract and Blood, and Its Activity Is Controlled by CcpA and GlnR.Front Cell Infect Microbiol. 2019 Sep 13;9:326. doi: 10.3389/fcimb.2019.00326. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31572692 Free PMC article.
-
Amino Acid Starvation-Induced Glutamine Accumulation Enhances Pneumococcal Survival.mSphere. 2023 Jun 22;8(3):e0062522. doi: 10.1128/msphere.00625-22. Epub 2023 Apr 5. mSphere. 2023. PMID: 37017541 Free PMC article.
-
Time-resolved transcriptomic and proteomic profiling of Heyndrickxia coagulans during NaOH-buffered L-lactic acid production.Front Microbiol. 2023 Nov 29;14:1296692. doi: 10.3389/fmicb.2023.1296692. eCollection 2023. Front Microbiol. 2023. PMID: 38094625 Free PMC article.
-
Role of the pyruvate metabolic network on carbohydrate metabolism and virulence in Streptococcus pneumoniae.Mol Microbiol. 2020 Oct;114(4):536-552. doi: 10.1111/mmi.14557. Epub 2020 Jun 24. Mol Microbiol. 2020. PMID: 32495474 Free PMC article.
References
-
- Kadioglu A., Weiser J., Paton J. & Andrew P. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6, 288–301 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

