Formation of Nitrogenase NifDK Tetramers in the Mitochondria of Saccharomyces cerevisiae
- PMID: 28221768
- PMCID: PMC5477005
- DOI: 10.1021/acssynbio.6b00371
Formation of Nitrogenase NifDK Tetramers in the Mitochondria of Saccharomyces cerevisiae
Abstract
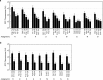
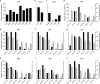
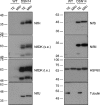
Transferring the prokaryotic enzyme nitrogenase into a eukaryotic host with the final aim of developing N2 fixing cereal crops would revolutionize agricultural systems worldwide. Targeting it to mitochondria has potential advantages because of the organelle's high O2 consumption and the presence of bacterial-type iron-sulfur cluster biosynthetic machinery. In this study, we constructed 96 strains of Saccharomyces cerevisiae in which transcriptional units comprising nine Azotobacter vinelandii nif genes (nifHDKUSMBEN) were integrated into the genome. Two combinatorial libraries of nif gene clusters were constructed: a library of mitochondrial leading sequences consisting of 24 clusters within four subsets of nif gene expression strength, and an expression library of 72 clusters with fixed mitochondrial leading sequences and nif expression levels assigned according to factorial design. In total, 29 promoters and 18 terminators were combined to adjust nif gene expression levels. Expression and mitochondrial targeting was confirmed at the protein level as immunoblot analysis showed that Nif proteins could be efficiently accumulated in mitochondria. NifDK tetramer formation, an essential step of nitrogenase assembly, was experimentally proven both in cell-free extracts and in purified NifDK preparations. This work represents a first step toward obtaining functional nitrogenase in the mitochondria of a eukaryotic cell.
Keywords: Azotobacter vinelandii; NifDK; mitochondria; nif genes; nitrogen fixation; yeast.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Biosynthesis of the nitrogenase active-site cofactor precursor NifB-co in Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2019 Dec 10;116(50):25078-25086. doi: 10.1073/pnas.1904903116. Epub 2019 Nov 25. Proc Natl Acad Sci U S A. 2019. PMID: 31767756 Free PMC article.
-
Construction of recombinant Escherichia coli producing nitrogenase-related proteins from Azotobacter vinelandii.Biosci Biotechnol Biochem. 2021 Sep 22;85(10):2209-2216. doi: 10.1093/bbb/zbab144. Biosci Biotechnol Biochem. 2021. PMID: 34387317
-
Combined Assembly and Targeted Integration of Multigene for Nitrogenase Biosynthetic Pathway in Saccharomyces cerevisiae.ACS Synth Biol. 2019 Aug 16;8(8):1766-1775. doi: 10.1021/acssynbio.9b00060. Epub 2019 Jul 22. ACS Synth Biol. 2019. PMID: 31117360
-
Challenges to develop nitrogen-fixing cereals by direct nif-gene transfer.Plant Sci. 2014 Aug;225:130-7. doi: 10.1016/j.plantsci.2014.06.003. Epub 2014 Jun 11. Plant Sci. 2014. PMID: 25017168 Review.
-
Molecular Mechanism and Agricultural Application of the NifA-NifL System for Nitrogen Fixation.Int J Mol Sci. 2023 Jan 4;24(2):907. doi: 10.3390/ijms24020907. Int J Mol Sci. 2023. PMID: 36674420 Free PMC article. Review.
Cited by
-
Nitrogen fixation in maize: breeding opportunities.Theor Appl Genet. 2021 May;134(5):1263-1280. doi: 10.1007/s00122-021-03791-5. Epub 2021 Mar 7. Theor Appl Genet. 2021. PMID: 33677701 Review.
-
Nitrogenase Cofactor Maturase NifB Isolated from Transgenic Rice is Active in FeMo-co Synthesis.ACS Synth Biol. 2022 Sep 16;11(9):3028-3036. doi: 10.1021/acssynbio.2c00194. Epub 2022 Aug 23. ACS Synth Biol. 2022. PMID: 35998307 Free PMC article.
-
Adaptation of the GoldenBraid modular cloning system and creation of a toolkit for the expression of heterologous proteins in yeast mitochondria.BMC Biotechnol. 2017 Nov 13;17(1):80. doi: 10.1186/s12896-017-0393-y. BMC Biotechnol. 2017. PMID: 29132331 Free PMC article.
-
Engineering Nitrogen Fixation Activity in an Oxygenic Phototroph.mBio. 2018 Jun 5;9(3):e01029-18. doi: 10.1128/mBio.01029-18. mBio. 2018. PMID: 29871920 Free PMC article.
-
Use of synthetic biology tools to optimize the production of active nitrogenase Fe protein in chloroplasts of tobacco leaf cells.Plant Biotechnol J. 2020 Sep;18(9):1882-1896. doi: 10.1111/pbi.13347. Epub 2020 Apr 7. Plant Biotechnol J. 2020. PMID: 31985876 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

