The acyl-activating enzyme PhAAE13 is an alternative enzymatic source of precursors for anthocyanin biosynthesis in petunia flowers
- PMID: 28204578
- PMCID: PMC5441920
- DOI: 10.1093/jxb/erw426
The acyl-activating enzyme PhAAE13 is an alternative enzymatic source of precursors for anthocyanin biosynthesis in petunia flowers
Abstract
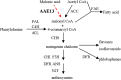
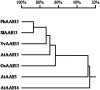
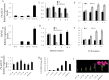
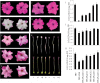
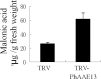
Anthocyanins, a class of flavonoids, are responsible for the orange to blue coloration of flowers and act as visual attractors to aid pollination and seed dispersal. Malonyl-CoA is the precursor for the formation of flavonoids and anthocyanins. Previous studies have suggested that malonyl-CoA is formed almost exclusively by acetyl-CoA carboxylase, which catalyzes the ATP-dependent formation of malonyl-CoA from acetyl-CoA and bicarbonate. In the present study, the full-length cDNA of Petunia hybrida acyl-activating enzyme 13 (PhAAE13), a member of clade VII of the AAE superfamily that encodes malonyl-CoA synthetase, was isolated. The expression of PhAAE13 was highest in corollas and was down-regulated by ethylene. Virus-induced gene silencing of petunia PhAAE13 significantly reduced anthocyanin accumulation, fatty acid content, and cuticular wax components content, and increased malonic acid content in flowers. The silencing of PhAAE3 and PhAAE14, the other two genes in clade VII of the AAE superfamily, did not change the anthocyanin content in petunia flowers. This study provides strong evidence indicating that PhAAE13, among clade VII of the AAE superfamily, is specifically involved in anthocyanin biosynthesis in petunia flowers.
Keywords: AAE13; anthocyanin synthesis; malonic acid; malonyl-CoA; petunia.
Figures

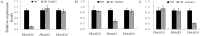
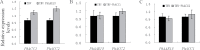
Similar articles
-
CCoAOMT Down-Regulation Activates Anthocyanin Biosynthesis in Petunia.Plant Physiol. 2016 Feb;170(2):717-31. doi: 10.1104/pp.15.01646. Epub 2015 Nov 30. Plant Physiol. 2016. PMID: 26620524 Free PMC article.
-
cDNA cloning, heterologous expressions, and functional characterization of malonyl-coenzyme a:anthocyanidin 3-o-glucoside-6"-o-malonyltransferase from dahlia flowers.Plant Physiol. 2002 Dec;130(4):2142-51. doi: 10.1104/pp.010447. Plant Physiol. 2002. PMID: 12481098 Free PMC article.
-
Mitochondrial citrate synthase plays important roles in anthocyanin synthesis in petunia.Plant Sci. 2021 Apr;305:110835. doi: 10.1016/j.plantsci.2021.110835. Epub 2021 Feb 5. Plant Sci. 2021. PMID: 33691969
-
Regulation of volatile benzenoid biosynthesis in petunia flowers.Trends Plant Sci. 2006 Jan;11(1):20-5. doi: 10.1016/j.tplants.2005.09.009. Epub 2005 Oct 12. Trends Plant Sci. 2006. PMID: 16226052 Review.
-
Achievements and perspectives in biochemistry concerning anthocyanin modification for blue flower coloration.Plant Cell Physiol. 2015 Jan;56(1):28-40. doi: 10.1093/pcp/pcu097. Epub 2014 Jul 10. Plant Cell Physiol. 2015. PMID: 25015943 Review.
Cited by
-
Transcriptome-wide m6A methylation in natural yellow leaf of Catalpa fargesii.Front Plant Sci. 2023 Jun 19;14:1167789. doi: 10.3389/fpls.2023.1167789. eCollection 2023. Front Plant Sci. 2023. PMID: 37404531 Free PMC article.
-
Genome-Wide Identification and Gene Expression Analysis of Acyl-Activating Enzymes Superfamily in Tomato (Solanum lycopersicum) Under Aluminum Stress.Front Plant Sci. 2021 Dec 2;12:754147. doi: 10.3389/fpls.2021.754147. eCollection 2021. Front Plant Sci. 2021. PMID: 34925406 Free PMC article.
-
The R2R3-MYB transcription factor EVER controls the emission of petunia floral volatiles by regulating epicuticular wax biosynthesis in the petal epidermis.Plant Cell. 2023 Dec 21;36(1):174-193. doi: 10.1093/plcell/koad251. Plant Cell. 2023. PMID: 37818992 Free PMC article.
-
PaACL silencing accelerates flower senescence and changes the proteome to maintain metabolic homeostasis in Petunia hybrida.J Exp Bot. 2020 Aug 6;71(16):4858-4876. doi: 10.1093/jxb/eraa208. J Exp Bot. 2020. PMID: 32364241 Free PMC article.
-
Isolation, Diversity, and Antimicrobial and Immunomodulatory Activities of Endophytic Actinobacteria From Tea Cultivars Zijuan and Yunkang-10 (Camellia sinensis var. assamica).Front Microbiol. 2018 Jun 18;9:1304. doi: 10.3389/fmicb.2018.01304. eCollection 2018. Front Microbiol. 2018. PMID: 29967601 Free PMC article.
References
-
- Abbruzzese A, Park MH, Folk JE. 1986. Deoxyhypusine hydroxylase from rat testis. Partial purification and characterization. Journal of Biological Chemistry 261, 3085–3089. - PubMed
-
- Belkebir A, Benhassaine-Kesri G. 2013. Sethoxydim treatment inhibits lipid metabolism and enhances the accumulation of anthocyanins in rape (Brassica napus L.) leaves. Pesticide Biochemistry and Physiology 107, 120–126. - PubMed
-
- Black PN, DiRusso CC. 2007. Yeast acyl-CoA synthetases at the crossroads of fatty acid metabolism and regulation. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1771, 286–298. - PubMed
-
- Black PN, Zhang Q, Weimar JD, DiRusso CC. 1997. Mutational analysis of a fatty acyl-coenzyme A synthetase signature motif identifies seven amino acid residues that modulate fatty acid substrate specificity. Journal of Biological Chemistry 272, 4896–4903. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

