3D genome structure modeling by Lorentzian objective function
- PMID: 28180292
- PMCID: PMC5430849
- DOI: 10.1093/nar/gkw1155
3D genome structure modeling by Lorentzian objective function
Abstract
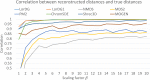
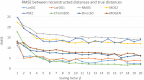
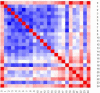
The 3D structure of the genome plays a vital role in biological processes such as gene interaction, gene regulation, DNA replication and genome methylation. Advanced chromosomal conformation capture techniques, such as Hi-C and tethered conformation capture, can generate chromosomal contact data that can be used to computationally reconstruct 3D structures of the genome. We developed a novel restraint-based method that is capable of reconstructing 3D genome structures utilizing both intra-and inter-chromosomal contact data. Our method was robust to noise and performed well in comparison with a panel of existing methods on a controlled simulated data set. On a real Hi-C data set of the human genome, our method produced chromosome and genome structures that are consistent with 3D FISH data and known knowledge about the human chromosome and genome, such as, chromosome territories and the cluster of small chromosomes in the nucleus center with the exception of the chromosome 18. The tool and experimental data are available at https://missouri.box.com/v/LorDG.
Figures
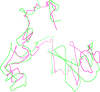




Similar articles
-
A maximum likelihood algorithm for reconstructing 3D structures of human chromosomes from chromosomal contact data.BMC Genomics. 2018 Feb 23;19(1):161. doi: 10.1186/s12864-018-4546-8. BMC Genomics. 2018. PMID: 29471801 Free PMC article.
-
Reconstructing spatial organizations of chromosomes through manifold learning.Nucleic Acids Res. 2018 May 4;46(8):e50. doi: 10.1093/nar/gky065. Nucleic Acids Res. 2018. PMID: 29408992 Free PMC article.
-
MOGEN: a tool for reconstructing 3D models of genomes from chromosomal conformation capturing data.Bioinformatics. 2016 May 1;32(9):1286-92. doi: 10.1093/bioinformatics/btv754. Epub 2015 Dec 31. Bioinformatics. 2016. PMID: 26722115
-
An Overview of Genome Organization and How We Got There: from FISH to Hi-C.Microbiol Mol Biol Rev. 2015 Sep;79(3):347-72. doi: 10.1128/MMBR.00006-15. Microbiol Mol Biol Rev. 2015. PMID: 26223848 Free PMC article. Review.
-
[Chromosome territories in the interphase nucleus in normal or pathological condition].Vestn Ross Akad Med Nauk. 2011;(9):48-54. Vestn Ross Akad Med Nauk. 2011. PMID: 22145372 Review. Russian.
Cited by
-
Application of Hi-C and other omics data analysis in human cancer and cell differentiation research.Comput Struct Biotechnol J. 2021 Apr 8;19:2070-2083. doi: 10.1016/j.csbj.2021.04.016. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33995903 Free PMC article. Review.
-
GSDB: a database of 3D chromosome and genome structures reconstructed from Hi-C data.BMC Mol Cell Biol. 2020 Aug 5;21(1):60. doi: 10.1186/s12860-020-00304-y. BMC Mol Cell Biol. 2020. PMID: 32758136 Free PMC article.
-
SCL: a lattice-based approach to infer 3D chromosome structures from single-cell Hi-C data.Bioinformatics. 2019 Oct 15;35(20):3981-3988. doi: 10.1093/bioinformatics/btz181. Bioinformatics. 2019. PMID: 30865261 Free PMC article.
-
Dynamic chromatin architectures provide insights into the genetics of cattle myogenesis.J Anim Sci Biotechnol. 2023 Apr 14;14(1):59. doi: 10.1186/s40104-023-00855-y. J Anim Sci Biotechnol. 2023. PMID: 37055796 Free PMC article.
-
A compendium and comparative epigenomics analysis of cis-regulatory elements in the pig genome.Nat Commun. 2021 Apr 13;12(1):2217. doi: 10.1038/s41467-021-22448-x. Nat Commun. 2021. PMID: 33850120 Free PMC article.
References
-
- Cremer T., Cremer C.. Rise, fall and resurrection of chromosome territories: a historical perspective. Part I. The rise of chromosome territories. Eur. J. Histochem. 2006; 50:161–176. - PubMed
-
- Edelmann P., Bornfleth H., Zink D., Cremer T., Cremer C.. Morphology and dynamics of chromosome territories in living cells. Biochim. Biophys. Acta. 2001; 1551:M29–M39. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

