Mechanisms of Regulation of the Chemokine-Receptor Network
- PMID: 28178200
- PMCID: PMC5343877
- DOI: 10.3390/ijms18020342
Mechanisms of Regulation of the Chemokine-Receptor Network
Abstract
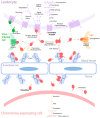
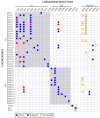
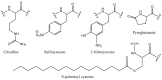
The interactions of chemokines with their G protein-coupled receptors promote the migration of leukocytes during normal immune function and as a key aspect of the inflammatory response to tissue injury or infection. This review summarizes the major cellular and biochemical mechanisms by which the interactions of chemokines with chemokine receptors are regulated, including: selective and competitive binding interactions; genetic polymorphisms; mRNA splice variation; variation of expression, degradation and localization; down-regulation by atypical (decoy) receptors; interactions with cell-surface glycosaminoglycans; post-translational modifications; oligomerization; alternative signaling responses; and binding to natural or pharmacological inhibitors.
Keywords: binding; chemokine; chemokine receptor; expression; glycosaminoglycan; inhibitor; oligomerization; post-translational modification; regulation; signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
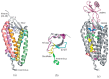
Similar articles
-
A guide to chemokines and their receptors.FEBS J. 2018 Aug;285(16):2944-2971. doi: 10.1111/febs.14466. Epub 2018 Apr 24. FEBS J. 2018. PMID: 29637711 Free PMC article. Review.
-
Chemokine cooperativity is caused by competitive glycosaminoglycan binding.J Immunol. 2014 Apr 15;192(8):3908-3914. doi: 10.4049/jimmunol.1302159. Epub 2014 Mar 17. J Immunol. 2014. PMID: 24639348 Free PMC article.
-
Chemokine oligomerization in cell signaling and migration.Prog Mol Biol Transl Sci. 2013;117:531-78. doi: 10.1016/B978-0-12-386931-9.00020-9. Prog Mol Biol Transl Sci. 2013. PMID: 23663982 Free PMC article. Review.
-
New insights into the structure and function of chemokine receptor:chemokine complexes from an experimental perspective.J Leukoc Biol. 2020 Jun;107(6):1115-1122. doi: 10.1002/JLB.2MR1219-288R. Epub 2020 Jan 22. J Leukoc Biol. 2020. PMID: 31965639 Review.
-
Differential structural remodelling of heparan sulfate by chemokines: the role of chemokine oligomerization.Open Biol. 2017 Jan;7(1):160286. doi: 10.1098/rsob.160286. Open Biol. 2017. PMID: 28123055 Free PMC article.
Cited by
-
Chemokine Receptors CCR1 and CCR2 on Peripheral Blood Mononuclear Cells of Newly Diagnosed Patients with the CD38-Positive Chronic Lymphocytic Leukemia.J Clin Med. 2020 Jul 21;9(7):2312. doi: 10.3390/jcm9072312. J Clin Med. 2020. PMID: 32708233 Free PMC article.
-
The influence of 4G/5G polymorphism in the plasminogen-activator-inhibitor-1 promoter on COVID-19 severity and endothelial dysfunction.Front Immunol. 2024 Aug 30;15:1445294. doi: 10.3389/fimmu.2024.1445294. eCollection 2024. Front Immunol. 2024. PMID: 39281671 Free PMC article.
-
Biased Signaling of CCL21 and CCL19 Does Not Rely on N-Terminal Differences, but Markedly on the Chemokine Core Domains and Extracellular Loop 2 of CCR7.Front Immunol. 2019 Sep 13;10:2156. doi: 10.3389/fimmu.2019.02156. eCollection 2019. Front Immunol. 2019. PMID: 31572374 Free PMC article.
-
Role of MCP-1 and CCR2 in alcohol neurotoxicity.Pharmacol Res. 2019 Jan;139:360-366. doi: 10.1016/j.phrs.2018.11.030. Epub 2018 Nov 22. Pharmacol Res. 2019. PMID: 30472461 Free PMC article. Review.
-
Evasins: Tick Salivary Proteins that Inhibit Mammalian Chemokines.Trends Biochem Sci. 2020 Feb;45(2):108-122. doi: 10.1016/j.tibs.2019.10.003. Epub 2019 Nov 1. Trends Biochem Sci. 2020. PMID: 31679840 Free PMC article. Review.
References
-
- Dimberg A. Chemokines in angiogenesis. Curr. Top. Microbiol. Immunol. 2010;341:59–80. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

