Linker histones: novel insights into structure-specific recognition of the nucleosome
- PMID: 28177778
- PMCID: PMC5654525
- DOI: 10.1139/bcb-2016-0097
Linker histones: novel insights into structure-specific recognition of the nucleosome
Abstract
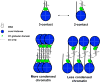
Linker histones (H1s) are a primary component of metazoan chromatin, fulfilling numerous functions, both in vitro and in vivo, including stabilizing the wrapping of DNA around the nucleosome, promoting folding and assembly of higher order chromatin structures, influencing nucleosome spacing on DNA, and regulating specific gene expression. However, many molecular details of how H1 binds to nucleosomes and recognizes unique structural features on the nucleosome surface remain undefined. Numerous, confounding studies are complicated not only by experimental limitations, but the use of different linker histone isoforms and nucleosome constructions. This review summarizes the decades of research that has resulted in several models of H1 association with nucleosomes, with a focus on recent advances that suggest multiple modes of H1 interaction in chromatin, while highlighting the remaining questions.
Keywords: ADN; DNA; histone de liaison; linker histone; nucleosome; nucléosome.
Figures

Similar articles
-
Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1.Mol Cell. 2017 May 4;66(3):384-397.e8. doi: 10.1016/j.molcel.2017.04.012. Mol Cell. 2017. PMID: 28475873 Free PMC article.
-
HMGN1 and 2 remodel core and linker histone tail domains within chromatin.Nucleic Acids Res. 2017 Sep 29;45(17):9917-9930. doi: 10.1093/nar/gkx579. Nucleic Acids Res. 2017. PMID: 28973435 Free PMC article.
-
Emerging roles of linker histones in regulating chromatin structure and function.Nat Rev Mol Cell Biol. 2018 Mar;19(3):192-206. doi: 10.1038/nrm.2017.94. Epub 2017 Oct 11. Nat Rev Mol Cell Biol. 2018. PMID: 29018282 Free PMC article. Review.
-
Binding Dynamics of Disordered Linker Histone H1 with a Nucleosomal Particle.J Mol Biol. 2021 Mar 19;433(6):166881. doi: 10.1016/j.jmb.2021.166881. Epub 2021 Feb 20. J Mol Biol. 2021. PMID: 33617899 Free PMC article.
-
Chromatin structures condensed by linker histones.Essays Biochem. 2019 Apr 23;63(1):75-87. doi: 10.1042/EBC20180056. Print 2019 Apr 23. Essays Biochem. 2019. PMID: 31015384 Review.
Cited by
-
Regulation of chromatin architecture by protein binding: insights from molecular modeling.Biophys Rev. 2024 May 9;16(3):331-343. doi: 10.1007/s12551-024-01195-5. eCollection 2024 Jun. Biophys Rev. 2024. PMID: 39099845 Review.
-
Linker histone epitopes are hidden by in situ higher-order chromatin structure.Epigenetics Chromatin. 2020 Jun 6;13(1):26. doi: 10.1186/s13072-020-00345-9. Epigenetics Chromatin. 2020. PMID: 32505195 Free PMC article.
-
Structure and function of archaeal histones.PLoS Genet. 2018 Sep 13;14(9):e1007582. doi: 10.1371/journal.pgen.1007582. eCollection 2018 Sep. PLoS Genet. 2018. PMID: 30212449 Free PMC article. Review.
-
Targeting Chromatin Remodeling for Cancer Therapy.Curr Mol Pharmacol. 2019;12(3):215-229. doi: 10.2174/1874467212666190215112915. Curr Mol Pharmacol. 2019. PMID: 30767757 Free PMC article. Review.
-
The Role of PARP1 and PAR in ATP-Independent Nucleosome Reorganisation during the DNA Damage Response.Genes (Basel). 2022 Dec 30;14(1):112. doi: 10.3390/genes14010112. Genes (Basel). 2022. PMID: 36672853 Free PMC article. Review.
References
-
- Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. - PubMed
-
- Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. Journal of molecular biology. 1986;187:591–601. - PubMed
-
- Bohm L, Mitchell TC. Sequence conservation in the N-terminal domain of histone H1. FEBS letters. 1985;193:1–4. - PubMed
-
- Bradbury EM, Chapman GE, Danby SE, Hartman PG, Riches PL. Studies on the role and mode of operation of the very-lysine-rich histone H1 (F1) in eukaryote chromatin. The properties of the N-terminal and C-terminal halves of histone H1. European journal of biochemistry / FEBS. 1975;57:521–528. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

