Keep it together: restraints in crystallographic refinement of macromolecule-ligand complexes
- PMID: 28177305
- PMCID: PMC5297912
- DOI: 10.1107/S2059798316017964
Keep it together: restraints in crystallographic refinement of macromolecule-ligand complexes
Abstract
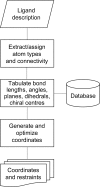
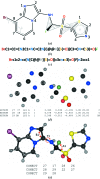
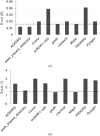
A short introduction is provided to the concept of restraints in macromolecular crystallographic refinement. A typical ligand restraint-generation process is then described, covering types of input, the methodology and the mechanics behind the software in general terms, how this has evolved over recent years and what to look for in the output. Finally, the currently available restraint-generation software is compared, concluding with some thoughts for the future.
Keywords: ligand complexes; macromolecular crystallography; refinement; restraint sets; standard deviation.
Figures


Similar articles
-
In situ ligand restraints from quantum-mechanical methods.Acta Crystallogr D Struct Biol. 2023 Feb 1;79(Pt 2):100-110. doi: 10.1107/S2059798323000025. Epub 2023 Jan 20. Acta Crystallogr D Struct Biol. 2023. PMID: 36762856 Free PMC article.
-
High-throughput quantum-mechanics/molecular-mechanics (ONIOM) macromolecular crystallographic refinement with PHENIX/DivCon: the impact of mixed Hamiltonian methods on ligand and protein structure.Acta Crystallogr D Struct Biol. 2018 Nov 1;74(Pt 11):1063-1077. doi: 10.1107/S2059798318012913. Epub 2018 Oct 29. Acta Crystallogr D Struct Biol. 2018. PMID: 30387765 Free PMC article.
-
Accurate macromolecular crystallographic refinement: incorporation of the linear scaling, semiempirical quantum-mechanics program DivCon into the PHENIX refinement package.Acta Crystallogr D Biol Crystallogr. 2014 May;70(Pt 5):1233-47. doi: 10.1107/S1399004714002260. Epub 2014 Apr 26. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24816093 Free PMC article.
-
Pound-wise but penny-foolish: How well do micromolecules fare in macromolecular refinement?Structure. 2003 Sep;11(9):1051-9. doi: 10.1016/s0969-2126(03)00186-2. Structure. 2003. PMID: 12962624 Review.
-
Recent developments for crystallographic refinement of macromolecules.Methods Mol Biol. 1996;56:245-66. doi: 10.1385/0-89603-259-0:245. Methods Mol Biol. 1996. PMID: 8781249 Review. No abstract available.
Cited by
-
Ligand fitting with CCP4.Acta Crystallogr D Struct Biol. 2017 Feb 1;73(Pt 2):158-170. doi: 10.1107/S2059798316020143. Epub 2017 Feb 1. Acta Crystallogr D Struct Biol. 2017. PMID: 28177312 Free PMC article.
-
On modelling disordered crystal structures through restraints from molecule-in-cluster computations, and distinguishing static and dynamic disorder.IUCrJ. 2021 Feb 18;8(Pt 2):305-318. doi: 10.1107/S2052252521000531. eCollection 2021 Mar 1. IUCrJ. 2021. PMID: 33708406 Free PMC article.
-
In situ ligand restraints from quantum-mechanical methods.Acta Crystallogr D Struct Biol. 2023 Feb 1;79(Pt 2):100-110. doi: 10.1107/S2059798323000025. Epub 2023 Jan 20. Acta Crystallogr D Struct Biol. 2023. PMID: 36762856 Free PMC article.
-
Refining the macromolecular model - achieving the best agreement with the data from X-ray diffraction experiment.Crystallogr Rev. 2018;24(4):236-262. doi: 10.1080/0889311X.2018.1521805. Epub 2018 Sep 21. Crystallogr Rev. 2018. PMID: 30416256 Free PMC article.
-
The missing link: covalent linkages in structural models.Acta Crystallogr D Struct Biol. 2021 Jun 1;77(Pt 6):727-745. doi: 10.1107/S2059798321003934. Epub 2021 May 19. Acta Crystallogr D Struct Biol. 2021. PMID: 34076588 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

