Direct Visualization of RNA-DNA Primer Removal from Okazaki Fragments Provides Support for Flap Cleavage and Exonucleolytic Pathways in Eukaryotic Cells
- PMID: 28159842
- PMCID: PMC5377794
- DOI: 10.1074/jbc.M116.758599
Direct Visualization of RNA-DNA Primer Removal from Okazaki Fragments Provides Support for Flap Cleavage and Exonucleolytic Pathways in Eukaryotic Cells
Abstract
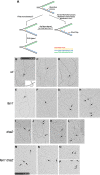
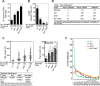
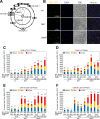
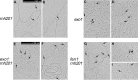
During DNA replication in eukaryotic cells, short single-stranded DNA segments known as Okazaki fragments are first synthesized on the lagging strand. The Okazaki fragments originate from ∼35-nucleotide-long RNA-DNA primers. After Okazaki fragment synthesis, these primers must be removed to allow fragment joining into a continuous lagging strand. To date, the models of enzymatic machinery that removes the RNA-DNA primers have come almost exclusively from biochemical reconstitution studies and some genetic interaction assays, and there is little direct evidence to confirm these models. One obstacle to elucidating Okazaki fragment processing has been the lack of methods that can directly examine primer removal in vivo In this study, we developed an electron microscopy assay that can visualize nucleotide flap structures on DNA replication forks in fission yeast (Schizosaccharomyces pombe). With this assay, we first demonstrated the generation of flap structures during Okazaki fragment processing in vivo The mean and median lengths of the flaps in wild-type cells were ∼51 and ∼41 nucleotides, respectively. We also used yeast mutants to investigate the impact of deleting key DNA replication nucleases on these flap structures. Our results provided direct in vivo evidence for a previously proposed flap cleavage pathway and the critical function of Dna2 and Fen1 in cleaving these flaps. In addition, we found evidence for another previously proposed exonucleolytic pathway involving RNA-DNA primer digestion by exonucleases RNase H2 and Exo1. Taken together, our observations suggest a dual mechanism for Okazaki fragment maturation in lagging strand synthesis and establish a new strategy for interrogation of this fascinating process.
Keywords: DNA enzyme; DNA replication; DNA structure; Dna2; Exo1; Fen1; Okazaki fragment processing; deoxyribonuclease (DNase); endonuclease; flap structures.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

Similar articles
-
Significance of the dissociation of Dna2 by flap endonuclease 1 to Okazaki fragment processing in Saccharomyces cerevisiae.J Biol Chem. 2009 Mar 27;284(13):8283-91. doi: 10.1074/jbc.M809189200. Epub 2009 Jan 29. J Biol Chem. 2009. PMID: 19179330 Free PMC article.
-
Reconstituted Okazaki fragment processing indicates two pathways of primer removal.J Biol Chem. 2006 Sep 8;281(36):26051-61. doi: 10.1074/jbc.M604805200. Epub 2006 Jul 11. J Biol Chem. 2006. PMID: 16837458
-
Dynamics of enzymatic interactions during short flap human Okazaki fragment processing by two forms of human DNA polymerase δ.DNA Repair (Amst). 2013 Nov;12(11):922-35. doi: 10.1016/j.dnarep.2013.08.008. Epub 2013 Sep 10. DNA Repair (Amst). 2013. PMID: 24035200 Free PMC article.
-
Okazaki fragment maturation: nucleases take centre stage.J Mol Cell Biol. 2011 Feb;3(1):23-30. doi: 10.1093/jmcb/mjq048. J Mol Cell Biol. 2011. PMID: 21278448 Free PMC article. Review.
-
Mechanism of Lagging-Strand DNA Replication in Eukaryotes.Adv Exp Med Biol. 2017;1042:117-133. doi: 10.1007/978-981-10-6955-0_6. Adv Exp Med Biol. 2017. PMID: 29357056 Review.
Cited by
-
Specificity of end resection pathways for double-strand break regions containing ribonucleotides and base lesions.Nat Commun. 2020 Jun 18;11(1):3088. doi: 10.1038/s41467-020-16903-4. Nat Commun. 2020. PMID: 32555206 Free PMC article.
-
Processing of matched and mismatched rNMPs in DNA by archaeal ribonucleotide excision repair.iScience. 2023 Nov 17;26(12):108479. doi: 10.1016/j.isci.2023.108479. eCollection 2023 Dec 15. iScience. 2023. PMID: 38077150 Free PMC article.
-
Multiple roles of DNA2 nuclease/helicase in DNA metabolism, genome stability and human diseases.Nucleic Acids Res. 2020 Jan 10;48(1):16-35. doi: 10.1093/nar/gkz1101. Nucleic Acids Res. 2020. PMID: 31754720 Free PMC article. Review.
-
Mechanistic investigation of human maturation of Okazaki fragments reveals slow kinetics.Nat Commun. 2022 Nov 15;13(1):6973. doi: 10.1038/s41467-022-34751-2. Nat Commun. 2022. PMID: 36379932 Free PMC article.
-
Ribonuclease H2 Subunit A Preserves Genomic Integrity and Promotes Prostate Cancer Progression.Cancer Res Commun. 2022 Aug 25;2(8):870-883. doi: 10.1158/2767-9764.CRC-22-0126. eCollection 2022 Aug. Cancer Res Commun. 2022. PMID: 36923313 Free PMC article.
References
-
- Sakabe K., and Okazaki R. (1966) A unique property of the replicating region of chromosomal DNA. Biochim. Biophys. Acta 129, 651–654 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

