The molecular basis of talin2's high affinity toward β1-integrin
- PMID: 28155884
- PMCID: PMC5290461
- DOI: 10.1038/srep41989
The molecular basis of talin2's high affinity toward β1-integrin
Abstract
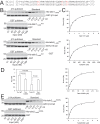
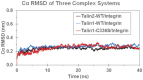
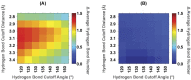
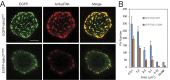
Talin interacts with β-integrin tails and actin to control integrin activation, thus regulating focal adhesion dynamics and cell migration. There are two talin genes, Tln1 and Tln2, which encode talin1 and talin2, and it is generally believed that talin2 functions redundantly with talin1. However, we show here that talin2 has a higher affinity to β1-integrin tails than talin1. Mutation of talin2 S339 to leucine, which can cause Fifth Finger Camptodactyly, a human genetic disease, completely disrupted its binding to β-integrin tails. Also, substitution of talin1 C336 with Ser enhanced the affinity of talin1, whereas substitution of talin2 S339 with Cys diminished that of talin2. Further computational modeling analysis shows that talin2 S339 formed a hydrogen bond with E353, which is critical for inducing key hydrogen bonds between talin2 N326 and β1-integrin R760, and between talin2 K327 and β1-integrin D759. Mutation at any of these residues significantly diminished the interaction of talin2 with β1- integrin tails. These hydrogen bonds were not observed in talin1/β1-integrin, but did exist in talin1C336S/β1-integrin complex. These results suggest that talin2 S339 forms a hydrogen bond with E353 to mediate its high affinity to β1-integrin.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Talin2-mediated traction force drives matrix degradation and cell invasion.J Cell Sci. 2016 Oct 1;129(19):3661-3674. doi: 10.1242/jcs.185959. J Cell Sci. 2016. PMID: 27694340 Free PMC article.
-
Roles of Talin2 in Traction Force Generation, Tumor Metastasis and Cardiovascular Integrity.Curr Protein Pept Sci. 2018;19(11):1071-1078. doi: 10.2174/1389203719666180809094731. Curr Protein Pept Sci. 2018. PMID: 30091412 Review.
-
Structural diversity in integrin/talin interactions.Structure. 2010 Dec 8;18(12):1654-66. doi: 10.1016/j.str.2010.09.018. Structure. 2010. PMID: 21134644 Free PMC article.
-
Talin1 phosphorylation activates β1 integrins: a novel mechanism to promote prostate cancer bone metastasis.Oncogene. 2015 Apr 2;34(14):1811-21. doi: 10.1038/onc.2014.116. Epub 2014 May 5. Oncogene. 2015. PMID: 24793790 Free PMC article.
-
The tale of two talins - two isoforms to fine-tune integrin signalling.FEBS Lett. 2018 Jun;592(12):2108-2125. doi: 10.1002/1873-3468.13081. Epub 2018 May 18. FEBS Lett. 2018. PMID: 29723415 Free PMC article. Review.
Cited by
-
PtdIns(4,5)P2 and PtdIns(3,4,5)P3 dynamics during focal adhesions assembly and disassembly in a cancer cell line.Turk J Biol. 2020 Dec 14;44(6):381-392. doi: 10.3906/biy-2004-108. eCollection 2020. Turk J Biol. 2020. PMID: 33402865 Free PMC article.
-
Synergistic Beneficial Effect of Docosahexaenoic Acid (DHA) and Docetaxel on the Expression Level of Matrix Metalloproteinase-2 (MMP-2) and MicroRNA-106b in Gastric Cancer.J Gastrointest Cancer. 2020 Mar;51(1):70-75. doi: 10.1007/s12029-019-00205-0. J Gastrointest Cancer. 2020. PMID: 30680612
-
A possible molecular mechanism for mechanotransduction at cellular focal adhesion complexes.Biophys Rep (N Y). 2021 Jul 21;1(1):100006. doi: 10.1016/j.bpr.2021.100006. eCollection 2021 Sep 8. Biophys Rep (N Y). 2021. PMID: 36425310 Free PMC article.
-
Talin2 binds to non-muscle myosin IIa and regulates cell attachment and fibronectin secretion.Sci Rep. 2024 Aug 30;14(1):20175. doi: 10.1038/s41598-024-70866-w. Sci Rep. 2024. PMID: 39215026 Free PMC article.
-
The role of talin2 in breast cancer tumorigenesis and metastasis.Oncotarget. 2017 Nov 6;8(63):106876-106887. doi: 10.18632/oncotarget.22449. eCollection 2017 Dec 5. Oncotarget. 2017. PMID: 29290996 Free PMC article.
References
-
- Hynes R. O. Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 (2002). - PubMed
-
- Scales T. M. E. & Parsons M. Spatial and temporal regulation of integrin signalling during cell migration. Curr Opin Cell Biol 23, 562–568 (2011). - PubMed
-
- Missan D. S. & DiPersio M. Integrin Control of Tumor Invasion. Crit Rev Eukaryot Gene Expr 22, 309–324 (2012). - PubMed
-
- Mercurio A. M. & Rabinovitz I. Towards a mechanistic understanding of tumor invasion—lessons from theα 6 β 4integrin. Semin Cancer Biol 11, 129–141 (2001). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

