Genetic and epigenetic control of the spatial organization of the genome
- PMID: 28137949
- PMCID: PMC5341720
- DOI: 10.1091/mbc.E16-03-0149
Genetic and epigenetic control of the spatial organization of the genome
Abstract
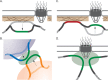
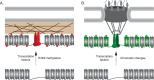
Eukaryotic genomes are spatially organized within the nucleus by chromosome folding, interchromosomal contacts, and interaction with nuclear structures. This spatial organization is observed in diverse organisms and both reflects and contributes to gene expression and differentiation. This leads to the notion that the arrangement of the genome within the nucleus has been shaped and conserved through evolutionary processes and likely plays an adaptive function. Both DNA-binding proteins and changes in chromatin structure influence the positioning of genes and larger domains within the nucleus. This suggests that the spatial organization of the genome can be genetically encoded by binding sites for DNA-binding proteins and can also involve changes in chromatin structure, potentially through nongenetic mechanisms. Here I briefly discuss the results that support these ideas and their implications for how genomes encode spatial organization.
© 2017 Brickner. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures
Similar articles
-
Beyond the sequence: cellular organization of genome function.Cell. 2007 Feb 23;128(4):787-800. doi: 10.1016/j.cell.2007.01.028. Cell. 2007. PMID: 17320514 Review.
-
Structural-Functional Domains of the Eukaryotic Genome.Biochemistry (Mosc). 2018 Apr;83(4):302-312. doi: 10.1134/S0006297918040028. Biochemistry (Mosc). 2018. PMID: 29626918 Review.
-
The genome in space and time: does form always follow function? How does the spatial and temporal organization of a eukaryotic genome reflect and influence its functions?Bioessays. 2012 Sep;34(9):800-10. doi: 10.1002/bies.201200034. Epub 2012 Jul 6. Bioessays. 2012. PMID: 22777837 Free PMC article.
-
Gene Positioning Effects on Expression in Eukaryotes.Annu Rev Genet. 2015;49:627-46. doi: 10.1146/annurev-genet-112414-055008. Epub 2015 Oct 2. Annu Rev Genet. 2015. PMID: 26436457 Review.
-
Epigenomics in 3D: importance of long-range spreading and specific interactions in epigenomic maintenance.Nucleic Acids Res. 2018 Mar 16;46(5):2252-2264. doi: 10.1093/nar/gky009. Nucleic Acids Res. 2018. PMID: 29365171 Free PMC article.
Cited by
-
Modeling homologous chromosome recognition via nonspecific interactions.Proc Natl Acad Sci U S A. 2024 May 14;121(20):e2317373121. doi: 10.1073/pnas.2317373121. Epub 2024 May 9. Proc Natl Acad Sci U S A. 2024. PMID: 38722810 Free PMC article.
-
Chromosome Territories in Hematological Malignancies.Cells. 2022 Apr 17;11(8):1368. doi: 10.3390/cells11081368. Cells. 2022. PMID: 35456046 Free PMC article. Review.
-
Brr6 plays a role in gene recruitment and transcriptional regulation at the nuclear envelope.Mol Biol Cell. 2018 Oct 15;29(21):2578-2590. doi: 10.1091/mbc.E18-04-0258. Epub 2018 Aug 22. Mol Biol Cell. 2018. PMID: 30133335 Free PMC article.
-
Nuclear Pore Proteins in Regulation of Chromatin State.Cells. 2019 Nov 9;8(11):1414. doi: 10.3390/cells8111414. Cells. 2019. PMID: 31717499 Free PMC article. Review.
-
Coaching from the sidelines: the nuclear periphery in genome regulation.Nat Rev Genet. 2019 Jan;20(1):39-50. doi: 10.1038/s41576-018-0063-5. Nat Rev Genet. 2019. PMID: 30356165 Free PMC article. Review.
References
-
- Apostolou E, Thanos D. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96. - PubMed
-
- Boveri T. Die blastermerenkerne von Ascaris megalocephala und die theorie der chromosomenindividualität. Arch Zellforsch. 1909;3:181.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

