An in vitro fluorescence based study of initiation of RNA synthesis by influenza B polymerase
- PMID: 28126917
- PMCID: PMC5399792
- DOI: 10.1093/nar/gkx043
An in vitro fluorescence based study of initiation of RNA synthesis by influenza B polymerase
Abstract
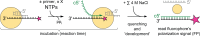
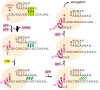
Influenza polymerase replicates, via a complementary RNA intermediate (cRNA), and transcribes the eight viral RNA (vRNA) genome segments. To initiate RNA synthesis it is bound to the conserved 5΄ and 3΄ extremities of the vRNA or cRNA (the 'promoter'). 5΄-3΄ base-pairing in the distal promoter region is essential to position the template RNA at the polymerase active site, as shown by a new crystal structure with the 3΄ end threading through the template entry tunnel. We develop fluorescence polarization assays to quantify initiation of cap-primed (transcription) or unprimed (replication) RNA synthesis by recombinant influenza B polymerase bound to the vRNA or cRNA promoter. The rate-limiting step is formation of a primed initiation complex with minimally ApG required to stabilize the 3΄ end of the template within the active-site. Polymerase bound to the vRNA promoter initiates RNA synthesis terminally, while the cRNA promoter directs internal initiation at a significantly lower rate. Progression to elongation requires breaking the promoter 5΄-3΄ base-pairing region and favourable compensation by the emerging template-product base-pairs. The RNA synthesis assay is adaptable to high-throughput screening for polymerase inhibitors. In a pilot study, we find that initiation at the cRNA promoter is unusually susceptible to inhibition by 2΄F-2΄dNTPs.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

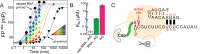



Similar articles
-
Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication.J Virol. 2006 Mar;80(5):2337-48. doi: 10.1128/JVI.80.5.2337-2348.2006. J Virol. 2006. PMID: 16474140 Free PMC article.
-
Mutation of an Influenza Virus Polymerase 3' RNA Promoter Binding Site Inhibits Transcription Elongation.J Virol. 2020 Jun 16;94(13):e00498-20. doi: 10.1128/JVI.00498-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32295915 Free PMC article.
-
Differential role of the influenza A virus polymerase PA subunit for vRNA and cRNA promoter binding.Virology. 2008 Jan 5;370(1):194-204. doi: 10.1016/j.virol.2007.08.029. Epub 2007 Oct 1. Virology. 2008. PMID: 17905403
-
Transcription and replication of the influenza a virus genome.Acta Virol. 2000 Oct;44(5):273-82. Acta Virol. 2000. PMID: 11252672 Review.
-
Structural insights into RNA synthesis by the influenza virus transcription-replication machine.Virus Res. 2017 Apr 15;234:103-117. doi: 10.1016/j.virusres.2017.01.013. Epub 2017 Jan 20. Virus Res. 2017. PMID: 28115197 Review.
Cited by
-
Initiation, Elongation, and Realignment during Influenza Virus mRNA Synthesis.J Virol. 2018 Jan 17;92(3):e01775-17. doi: 10.1128/JVI.01775-17. Print 2018 Feb 1. J Virol. 2018. PMID: 29142123 Free PMC article.
-
A novel mechanism of enhanced transcription activity and fidelity for influenza A viral RNA-dependent RNA polymerase.Nucleic Acids Res. 2021 Sep 7;49(15):8796-8810. doi: 10.1093/nar/gkab660. Nucleic Acids Res. 2021. PMID: 34379778 Free PMC article.
-
Structural snapshots of actively transcribing influenza polymerase.Nat Struct Mol Biol. 2019 Jun;26(6):460-470. doi: 10.1038/s41594-019-0232-z. Epub 2019 Jun 3. Nat Struct Mol Biol. 2019. PMID: 31160782 Free PMC article.
-
Identification of a Type-Specific Promoter Element That Differentiates between Influenza A and B Viruses.J Virol. 2019 Nov 13;93(23):e01164-19. doi: 10.1128/JVI.01164-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31534045 Free PMC article.
-
Biochemical characterization of the Lassa virus L protein.J Biol Chem. 2019 May 17;294(20):8088-8100. doi: 10.1074/jbc.RA118.006973. Epub 2019 Mar 29. J Biol Chem. 2019. PMID: 30926610 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

