5' to 3' mRNA Decay Contributes to the Regulation of Arabidopsis Seed Germination by Dormancy
- PMID: 28126845
- PMCID: PMC5338662
- DOI: 10.1104/pp.16.01933
5' to 3' mRNA Decay Contributes to the Regulation of Arabidopsis Seed Germination by Dormancy
Abstract
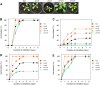
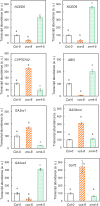
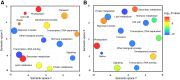
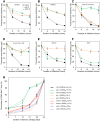
The regulation of plant gene expression, necessary for development and adaptive responses, relies not only on RNA transcription but also on messenger RNA (mRNA) fate. To understand whether seed germination relies on the degradation of specific subsets of mRNA, we investigated whether the 5' to 3' RNA decay machinery participated in the regulation of this process. Arabidopsis (Arabidopsis thaliana) seeds of exoribonuclease4 (xrn4) and varicose (vcs) mutants displayed distinct dormancy phenotypes. Transcriptome analysis of xrn4-5 and vcs-8 mutant seeds allowed us to identify genes that are likely to play a role in the control of germination. Study of 5' untranslated region features of these transcripts revealed that specific motifs, secondary energy, and GC content could play a role in their degradation by XRN4 and VCS, and Gene Ontology clustering revealed novel actors of seed dormancy and germination. Several specific transcripts identified as being putative targets of XRN4 and VCS in seeds (PECTIN LYASE-LIKE, ASPARTYL PROTEASE, DWD-HYPERSENSITIVE-TO-ABA3, and YELLOW STRIPE-LIKE5) were further studied by reverse genetics, and their functional roles in the germination process were confirmed by mutant analysis. These findings suggest that completion of germination and its regulation by dormancy also depend on the degradation of specific subsets of mRNA.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures

Similar articles
-
Germination Potential of Dormant and Nondormant Arabidopsis Seeds Is Driven by Distinct Recruitment of Messenger RNAs to Polysomes.Plant Physiol. 2015 Jul;168(3):1049-65. doi: 10.1104/pp.15.00510. Epub 2015 May 27. Plant Physiol. 2015. PMID: 26019300 Free PMC article.
-
AtPER1 enhances primary seed dormancy and reduces seed germination by suppressing the ABA catabolism and GA biosynthesis in Arabidopsis seeds.Plant J. 2020 Jan;101(2):310-323. doi: 10.1111/tpj.14542. Epub 2019 Oct 22. Plant J. 2020. PMID: 31536657
-
Combining association mapping and transcriptomics identify HD2B histone deacetylase as a genetic factor associated with seed dormancy in Arabidopsis thaliana.Plant J. 2013 Jun;74(5):815-28. doi: 10.1111/tpj.12167. Epub 2013 Apr 4. Plant J. 2013. PMID: 23464703
-
Parental and Environmental Control of Seed Dormancy in Arabidopsis thaliana.Annu Rev Plant Biol. 2022 May 20;73:355-378. doi: 10.1146/annurev-arplant-102820-090750. Epub 2022 Feb 9. Annu Rev Plant Biol. 2022. PMID: 35138879 Review.
-
From the Outside to the Inside: New Insights on the Main Factors That Guide Seed Dormancy and Germination.Genes (Basel). 2020 Dec 31;12(1):52. doi: 10.3390/genes12010052. Genes (Basel). 2020. PMID: 33396410 Free PMC article. Review.
Cited by
-
Biology in the Dry Seed: Transcriptome Changes Associated with Dry Seed Dormancy and Dormancy Loss in the Arabidopsis GA-Insensitive sleepy1-2 Mutant.Front Plant Sci. 2017 Dec 22;8:2158. doi: 10.3389/fpls.2017.02158. eCollection 2017. Front Plant Sci. 2017. PMID: 29312402 Free PMC article.
-
Computational and Experimental Tools to Monitor the Changes in Translation Efficiency of Plant mRNA on a Genome-Wide Scale: Advantages, Limitations, and Solutions.Int J Mol Sci. 2018 Dec 21;20(1):33. doi: 10.3390/ijms20010033. Int J Mol Sci. 2018. PMID: 30577638 Free PMC article. Review.
-
The role of RNA-binding protein, microRNA and alternative splicing in seed germination: a field need to be discovered.BMC Plant Biol. 2021 Apr 21;21(1):194. doi: 10.1186/s12870-021-02966-y. BMC Plant Biol. 2021. PMID: 33882821 Free PMC article. Review.
-
Analysis of Stored mRNA Degradation in Acceleratedly Aged Seeds of Wheat and Canola in Comparison to Arabidopsis.Plants (Basel). 2020 Dec 4;9(12):1707. doi: 10.3390/plants9121707. Plants (Basel). 2020. PMID: 33291562 Free PMC article.
-
SnRK2.10 kinase differentially modulates expression of hub WRKY transcription factors genes under salinity and oxidative stress in Arabidopsis thaliana.Front Plant Sci. 2023 Aug 9;14:1135240. doi: 10.3389/fpls.2023.1135240. eCollection 2023. Front Plant Sci. 2023. PMID: 37621885 Free PMC article.
References
-
- Bailly C, El-Maarouf-Bouteau H, Corbineau F (2008) From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. C R Biol 331: 806–814 - PubMed
-
- Bailly C, Leymarie J, Lehner A, Rousseau S, Côme D, Corbineau F (2004) Catalase activity and expression in developing sunflower seeds as related to drying. J Exp Bot 55: 475–483 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

