Development of a bimolecular luminescence complementation assay for RGS: G protein interactions in cells
- PMID: 28115169
- PMCID: PMC5330260
- DOI: 10.1016/j.ab.2017.01.013
Development of a bimolecular luminescence complementation assay for RGS: G protein interactions in cells
Abstract
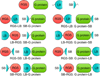
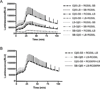
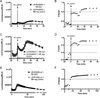
Cell based assessment tools and screening platforms are the preferred paradigm for small molecule identification and validation due to selectively identifying molecules with cellular activity and validation of compound activity against target proteins in their native environment. With respect to Regulator of G Protein Signaling (RGS) proteins, current cell based methodologies are either low throughput or monitor downstream signaling consequences. The increasing number of reports indicating RGS function in various disease pathogeneses highlights the need for a robust RGS inhibitor discovery and characterization paradigm. Promega's NanoBit Protein Complementation Assay utilizes NanoLuc, an engineered luciferase with enhanced luminescence characteristics which allow for both robust and kinetic assessment of protein interaction formation and disruption. Here we characterized 15 separate RGS: G protein interactions using this system. The binding profile of RGS: Gα interactions correlates to prior published biochemical binding profiles of these proteins. Additionally, we demonstrated this system is suitable for high throughput screening efforts via calculation of Z-factors for three of the interactions and demonstrated that a known small molecule inhibitor of RGS4 disrupts the RGS4: Gαi1 protein-protein interaction. In conclusion, the NanoBit Protein Complementation Assay holds promise as a robust platform for discovery and characterization of RGS inhibitors.
Keywords: Gα(i1); Gα(q); High throughput screening; NanoBit; RGS.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


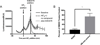
Similar articles
-
Interplay between negative and positive design elements in Gα helical domains of G proteins determines interaction specificity toward RGS2.Biochem J. 2018 Jul 26;475(14):2293-2304. doi: 10.1042/BCJ20180285. Biochem J. 2018. PMID: 29925530
-
RGS17/RGSZ2, a novel regulator of Gi/o, Gz, and Gq signaling.J Biol Chem. 2004 Jun 18;279(25):26314-22. doi: 10.1074/jbc.M401800200. Epub 2004 Apr 19. J Biol Chem. 2004. PMID: 15096504
-
RGS-insensitive G-protein mutations to study the role of endogenous RGS proteins.Methods Enzymol. 2004;389:229-43. doi: 10.1016/S0076-6879(04)89014-1. Methods Enzymol. 2004. PMID: 15313569 Review.
-
Gi and RGS proteins provide biochemical control of androgen receptor nuclear exclusion.J Mol Neurosci. 2007;31(1):1-12. doi: 10.1007/BF02686113. J Mol Neurosci. 2007. PMID: 17416965
-
Fluorescence-based assays for RGS box function.Methods Enzymol. 2004;389:56-71. doi: 10.1016/S0076-6879(04)89004-9. Methods Enzymol. 2004. PMID: 15313559 Review.
Cited by
-
Probing RAS Function Using Monobody and NanoBiT Technologies.Methods Mol Biol. 2024;2797:211-225. doi: 10.1007/978-1-0716-3822-4_15. Methods Mol Biol. 2024. PMID: 38570462 Free PMC article.
-
CcrZ is a pneumococcal spatiotemporal cell cycle regulator that interacts with FtsZ and controls DNA replication by modulating the activity of DnaA.Nat Microbiol. 2021 Sep;6(9):1175-1187. doi: 10.1038/s41564-021-00949-1. Epub 2021 Aug 9. Nat Microbiol. 2021. PMID: 34373624 Free PMC article.
-
Evaluation of the Selectivity and Cysteine Dependence of Inhibitors across the Regulator of G Protein-Signaling Family.Mol Pharmacol. 2018 Jan;93(1):25-35. doi: 10.1124/mol.117.109843. Epub 2017 Oct 19. Mol Pharmacol. 2018. PMID: 29051318 Free PMC article.
-
Lumit: A Homogeneous Bioluminescent Immunoassay for Detecting Diverse Analytes and Intracellular Protein Targets.Methods Mol Biol. 2023;2612:195-224. doi: 10.1007/978-1-0716-2903-1_15. Methods Mol Biol. 2023. PMID: 36795369
-
Luminescence- and Fluorescence-Based Complementation Assays to Screen for GPCR Oligomerization: Current State of the Art.Int J Mol Sci. 2019 Jun 17;20(12):2958. doi: 10.3390/ijms20122958. Int J Mol Sci. 2019. PMID: 31213021 Free PMC article. Review.
References
-
- Berman DM, Kozasa T, Gilman AG. The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J Biol Chem. 1996;271(44):27209–27212. - PubMed
-
- Blazer LL, Storaska AJ, Jutkiewicz EM, Turner EM, Calcagno M, Wade SM, Wang Q, Huang XP, Traynor JR, Husbands SM, Morari M, Neubig RR. Selectivity and anti-Parkinson's potential of thiadiazolidinone RGS4 inhibitors. ACS Chem Neurosci. 2015;6(6):911–919. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

