mTORC1 inhibition in cancer cells protects from glutaminolysis-mediated apoptosis during nutrient limitation
- PMID: 28112156
- PMCID: PMC5264013
- DOI: 10.1038/ncomms14124
mTORC1 inhibition in cancer cells protects from glutaminolysis-mediated apoptosis during nutrient limitation
Abstract
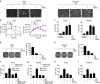
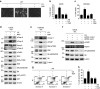
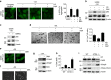
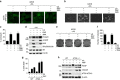
A master coordinator of cell growth, mTORC1 is activated by different metabolic inputs, particularly the metabolism of glutamine (glutaminolysis), to control a vast range of cellular processes, including autophagy. As a well-recognized tumour promoter, inhibitors of mTORC1 such as rapamycin have been approved as anti-cancer agents, but their overall outcome in patients is rather poor. Here we show that mTORC1 also presents tumour suppressor features in conditions of nutrient restrictions. Thus, the activation of mTORC1 by glutaminolysis during nutritional imbalance inhibits autophagy and induces apoptosis in cancer cells. Importantly, rapamycin treatment reactivates autophagy and prevents the mTORC1-mediated apoptosis. We also observe that the ability of mTORC1 to activate apoptosis is mediated by the adaptor protein p62. Thus, the mTORC1-mediated upregulation of p62 during nutrient imbalance induces the binding of p62 to caspase 8 and the subsequent activation of the caspase pathway. Our data highlight the role of autophagy as a survival mechanism upon rapamycin treatment.
Figures


Similar articles
-
Pharmacological inhibition of mTORC1 but not mTORC2 protects against human disc cellular apoptosis, senescence, and extracellular matrix catabolism through Akt and autophagy induction.Osteoarthritis Cartilage. 2019 Jun;27(6):965-976. doi: 10.1016/j.joca.2019.01.009. Epub 2019 Feb 1. Osteoarthritis Cartilage. 2019. PMID: 30716534
-
Selective interference of mTORC1/RAPTOR protects against human disc cellular apoptosis, senescence, and extracellular matrix catabolism with Akt and autophagy induction.Osteoarthritis Cartilage. 2017 Dec;25(12):2134-2146. doi: 10.1016/j.joca.2017.08.019. Epub 2017 Sep 6. Osteoarthritis Cartilage. 2017. PMID: 28888905
-
Glutamoptosis: A new cell death mechanism inhibited by autophagy during nutritional imbalance.Autophagy. 2017 Jun 3;13(6):1078-1079. doi: 10.1080/15548627.2017.1299315. Epub 2017 Mar 15. Autophagy. 2017. PMID: 28296535 Free PMC article.
-
The anticancer effects of curcumin via targeting the mammalian target of rapamycin complex 1 (mTORC1) signaling pathway.Pharmacol Res. 2020 Jun;156:104798. doi: 10.1016/j.phrs.2020.104798. Epub 2020 Apr 8. Pharmacol Res. 2020. PMID: 32278045 Review.
-
Glutaminolysis and autophagy in cancer.Autophagy. 2015;11(8):1198-208. doi: 10.1080/15548627.2015.1053680. Autophagy. 2015. PMID: 26054373 Free PMC article. Review.
Cited by
-
Metabolic resistance to the inhibition of mitochondrial transcription revealed by CRISPR-Cas9 screen.EMBO Rep. 2022 Jan 5;23(1):e53054. doi: 10.15252/embr.202153054. Epub 2021 Nov 15. EMBO Rep. 2022. PMID: 34779571 Free PMC article.
-
Escaping mTOR inhibition for cancer therapy: Tumor suppressor functions of mTOR.Mol Cell Oncol. 2017 Mar 3;4(3):e1297284. doi: 10.1080/23723556.2017.1297284. eCollection 2017. Mol Cell Oncol. 2017. PMID: 28616576 Free PMC article.
-
Downregulation of Glutamine Synthetase, not glutaminolysis, is responsible for glutamine addiction in Notch1-driven acute lymphoblastic leukemia.Mol Oncol. 2021 May;15(5):1412-1431. doi: 10.1002/1878-0261.12877. Epub 2021 Feb 13. Mol Oncol. 2021. PMID: 33314742 Free PMC article.
-
ELA/APELA precursor cleaved by furin displays tumor suppressor function in renal cell carcinoma through mTORC1 activation.JCI Insight. 2020 Jul 23;5(14):e129070. doi: 10.1172/jci.insight.129070. JCI Insight. 2020. PMID: 32516140 Free PMC article.
-
mTORC1 beyond anabolic metabolism: Regulation of cell death.J Cell Biol. 2022 Dec 5;221(12):e202208103. doi: 10.1083/jcb.202208103. Epub 2022 Oct 25. J Cell Biol. 2022. PMID: 36282248 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

