The roles of RRP15 in nucleolar formation, ribosome biogenesis and checkpoint control in human cells
- PMID: 28099941
- PMCID: PMC5355092
- DOI: 10.18632/oncotarget.14658
The roles of RRP15 in nucleolar formation, ribosome biogenesis and checkpoint control in human cells
Abstract
The nucleolus controls ribosome biogenesis and its perturbation induces nucleolar stress that inhibits cell cycle progression and activates checkpoint responses. Here, we investigate the roles of ribosomal RNA processing protein, RRP15, in nucleolar formation, ribosome biogenesis, cell cycle progression and checkpoint control in human cells. RRP15 is localized in the nucleolus and required for nucleolar formation. In contrast to the budding yeast Rrp15p that was reported as a component of pre-60S subunits, RRP15 is found in both pre-40S and pre-60S subunits and involved in regulating rRNA transcription and ribosome biogenesis. Perturbation of RRP15 induces nucleolar stress that activates RPL5/RPL11/5S rRNA (RP)-Mdm2-p53 axis checkpoint response and arrests cells at G1-G1/S in p53-proficient non-transformed RPE1 cells but not in p53-deficient HeLa and MCF7 tumor cells. Instead, p53-deficient HeLa and MCF7 cells with RRP15-dependent nucleolar stress enter S-phase with S-phase perturbation that activates ATR-Chk1- γH2AX axis DNA replication/damage checkpoint response, delaying S-G2/M progression and, ultimately, causing cell death. The selective checkpoint response, cell cycle inhibition and/or cytotoxicity induced by RRP15-dependent nucleolar stress in p53-proficient non-transformed cells and p53-deficient tumor cells suggest that RRP15 might be a potential target for cancer therapy.
Keywords: RRP15; checkpoint control; nucleolar stress; nucleolus; ribosome biogenesis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
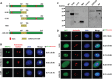
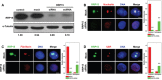
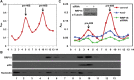
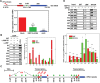


Similar articles
-
Perturbation of RNA Polymerase I transcription machinery by ablation of HEATR1 triggers the RPL5/RPL11-MDM2-p53 ribosome biogenesis stress checkpoint pathway in human cells.Cell Cycle. 2018;17(1):92-101. doi: 10.1080/15384101.2017.1403685. Epub 2017 Dec 10. Cell Cycle. 2018. PMID: 29143558 Free PMC article.
-
Acrolein preferentially damages nucleolus eliciting ribosomal stress and apoptosis in human cancer cells.Oncotarget. 2016 Dec 6;7(49):80450-80464. doi: 10.18632/oncotarget.12608. Oncotarget. 2016. PMID: 27741518 Free PMC article.
-
Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis.Biochim Biophys Acta. 2014 Jun;1842(6):817-30. doi: 10.1016/j.bbadis.2013.08.014. Epub 2013 Oct 26. Biochim Biophys Acta. 2014. PMID: 24514102 Review.
-
Functional ribosome biogenesis is a prerequisite for p53 destabilization: impact of chemotherapy on nucleolar functions and RNA metabolism.Biol Chem. 2013 Sep;394(9):1133-43. doi: 10.1515/hsz-2013-0153. Biol Chem. 2013. PMID: 23640940 Review.
-
Nucleolar stress: Molecular mechanisms and related human diseases.Cancer Sci. 2023 May;114(5):2078-2086. doi: 10.1111/cas.15755. Epub 2023 Feb 28. Cancer Sci. 2023. PMID: 36762786 Free PMC article. Review.
Cited by
-
Ribosomal Protein L15 is involved in Colon Carcinogenesis.Int J Med Sci. 2019 Aug 6;16(8):1132-1141. doi: 10.7150/ijms.34386. eCollection 2019. Int J Med Sci. 2019. PMID: 31523176 Free PMC article.
-
Different genetic mechanisms mediate spontaneous versus UVR-induced malignant melanoma.Elife. 2019 Jan 25;8:e42424. doi: 10.7554/eLife.42424. Elife. 2019. PMID: 30681412 Free PMC article.
-
Inhibition of Ribosomal RNA Processing 15 Homolog (RRP15) Suppressed Tumor Growth, Invasion and Epithelial to Mesenchymal Transition (EMT) of Colon Cancer.Int J Mol Sci. 2023 Feb 9;24(4):3528. doi: 10.3390/ijms24043528. Int J Mol Sci. 2023. PMID: 36834940 Free PMC article.
-
Morphological, physiological, and transcriptional responses of the freshwater diatom Fragilaria crotonensis to elevated pH conditions.Front Microbiol. 2022 Nov 25;13:1044464. doi: 10.3389/fmicb.2022.1044464. eCollection 2022. Front Microbiol. 2022. PMID: 36504786 Free PMC article.
-
Mapping the nucleolar proteome reveals a spatiotemporal organization related to intrinsic protein disorder.Mol Syst Biol. 2020 Aug;16(8):e9469. doi: 10.15252/msb.20209469. Mol Syst Biol. 2020. PMID: 32744794 Free PMC article.
References
-
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. - PubMed
-
- Orsolic I, Jurada D, Pullen N, Oren M, Eliopoulos AG, Volarevic S. The relationship between the nucleolus and cancer: Current evidence and emerging paradigms. Semin Cancer Biol. 2015 - PubMed
-
- Smirnov E, Cmarko D, Mazel T, Hornacek M, Raska I. Nucleolar DNA: the host and the guests. Histochem Cell Biol. 2016;145:359–372. - PubMed
-
- Nerurkar P, Altvater M, Gerhardy S, Schutz S, Fischer U, Weirich C, Panse VG. Eukaryotic Ribosome Assembly and Nuclear Export. Int Rev Cell Mol Biol. 2015;319:107–140. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

