Spread and Evolution of Respiratory Syncytial Virus A Genotype ON1, Coastal Kenya, 2010-2015
- PMID: 28098528
- PMCID: PMC5324789
- DOI: 10.3201/eid2302.161149
Spread and Evolution of Respiratory Syncytial Virus A Genotype ON1, Coastal Kenya, 2010-2015
Abstract
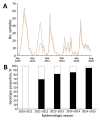
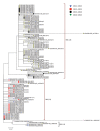
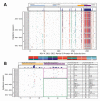
In February 2012, the novel respiratory syncytial virus (RSV) group A, genotype ON1, was detected in Kilifi County, coastal Kenya. ON1 is characterized by a 72-nt duplication within the highly variable G gene (encoding the immunogenic attachment surface protein). Cases were diagnosed through surveillance of pneumonia in children at the county hospital. Analysis of epidemiologic, clinical, and sequence data of RSV-A viruses detected over 5 RSV seasons (2010/2011 to 2014/2015) indicated the following: 1) replacement of previously circulating genotype GA2 ON1, 2) an abrupt expansion in the number of ON1 variants detected in the 2014/2015 epidemic, 3) recently accumulation of amino acid substitutions within the ON1 duplicated sequence, and 4) no clear evidence of altered pathogenicity relative to GA2. The study demonstrates the public health importance of molecular surveillance in defining the spread, clinical effects, and evolution of novel respiratory virus variants.
Keywords: G protein; RSV; evolutionary dynamics; genetic diversity; phylogenetic analysis; respiratory diseases; respiratory infections; respiratory syncytial virus; viruses.
Figures
Similar articles
-
Rapid spread and diversification of respiratory syncytial virus genotype ON1, Kenya.Emerg Infect Dis. 2014 Jun;20(6):950-9. doi: 10.3201/eid2006.131438. Emerg Infect Dis. 2014. PMID: 24856417 Free PMC article.
-
Molecular Evolutionary Dynamics of Respiratory Syncytial Virus Group A in Recurrent Epidemics in Coastal Kenya.J Virol. 2016 Apr 29;90(10):4990-5002. doi: 10.1128/JVI.03105-15. Print 2016 May 15. J Virol. 2016. PMID: 26937038 Free PMC article.
-
The genetic variability of glycoproteins among respiratory syncytial virus subtype A in China between 2009 and 2013.Infect Genet Evol. 2014 Oct;27:339-47. doi: 10.1016/j.meegid.2014.07.030. Epub 2014 Aug 7. Infect Genet Evol. 2014. PMID: 25109878
-
Respiratory Syncytial Virus G Protein Sequence Variability among Isolates from St. Petersburg, Russia, during the 2013-2014 Epidemic Season.Viruses. 2021 Jan 17;13(1):119. doi: 10.3390/v13010119. Viruses. 2021. PMID: 33477301 Free PMC article.
-
Rapid replacement of prevailing genotype of human respiratory syncytial virus by genotype ON1 in Beijing, 2012-2014.Infect Genet Evol. 2015 Jul;33:163-8. doi: 10.1016/j.meegid.2015.04.025. Epub 2015 Apr 27. Infect Genet Evol. 2015. PMID: 25929164
Cited by
-
Molecular epidemiology and characteristics of respiratory syncytial virus among hospitalized children in Guangzhou, China.Virol J. 2023 Nov 22;20(1):272. doi: 10.1186/s12985-023-02227-4. Virol J. 2023. PMID: 37993935 Free PMC article.
-
Molecular epidemiological surveillance of viral agents of acute lower respiratory tract infections in children in Accra, Ghana.BMC Pediatr. 2022 Jun 24;22(1):364. doi: 10.1186/s12887-022-03419-7. BMC Pediatr. 2022. PMID: 35751110 Free PMC article.
-
The significance of human respiratory syncytial virus (HRSV) in children from Ghana with acute lower respiratory tract infection: A molecular epidemiological analysis, 2006 and 2013-2014.PLoS One. 2018 Sep 10;13(9):e0203788. doi: 10.1371/journal.pone.0203788. eCollection 2018. PLoS One. 2018. PMID: 30199549 Free PMC article.
-
Circulation patterns and molecular epidemiology of human respiratory syncytial virus over five consecutive seasons in Morocco.Influenza Other Respir Viruses. 2023 Oct 17;17(10):e13203. doi: 10.1111/irv.13203. eCollection 2023 Oct. Influenza Other Respir Viruses. 2023. PMID: 37859975 Free PMC article.
-
Molecular epidemiology and clinical characteristics of respiratory syncytial virus in hospitalized children during winter 2021-2022 in Bengbu, China.Front Public Health. 2024 Jan 3;11:1310293. doi: 10.3389/fpubh.2023.1310293. eCollection 2023. Front Public Health. 2024. PMID: 38235154 Free PMC article.
References
-
- Agoti CN, Mwihuri AG, Sande CJ, Onyango CO, Medley GF, Cane PA, et al. Genetic relatedness of infecting and reinfecting respiratory syncytial virus strains identified in a birth cohort from rural Kenya. J Infect Dis. 2012;206:1532–41.http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&lis... 10.1093/infdis/jis570 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

