A comprehensive platform for the analysis of ubiquitin-like protein modifications using in vivo biotinylation
- PMID: 28098257
- PMCID: PMC5241687
- DOI: 10.1038/srep40756
A comprehensive platform for the analysis of ubiquitin-like protein modifications using in vivo biotinylation
Abstract
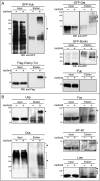
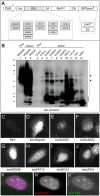
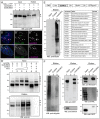
Post-translational modification by ubiquitin and ubiquitin-like proteins (UbLs) is fundamental for maintaining protein homeostasis. Efficient isolation of UbL conjugates is hampered by multiple factors, including cost and specificity of reagents, removal of UbLs by proteases, distinguishing UbL conjugates from interactors, and low quantities of modified substrates. Here we describe bioUbLs, a comprehensive set of tools for studying modifications in Drosophila and mammals, based on multicistronic expression and in vivo biotinylation using the E. coli biotin protein ligase BirA. While the bioUbLs allow rapid validation of UbL conjugation for exogenous or endogenous proteins, the single vector approach can facilitate biotinylation of most proteins of interest. Purification under denaturing conditions inactivates deconjugating enzymes and stringent washes remove UbL interactors and non-specific background. We demonstrate the utility of the method in Drosophila cells and transgenic flies, identifying an extensive set of putative SUMOylated proteins in both cases. For mammalian cells, we show conjugation and localization for many different UbLs, with the identification of novel potential substrates for UFM1. Ease of use and the flexibility to modify existing vectors will make the bioUbL system a powerful complement to existing strategies for studying this important mode of protein regulation.
Figures



Similar articles
-
Analysis of SUMOylated Proteins in Cells and In Vivo Using the bioSUMO Strategy.Methods Mol Biol. 2016;1475:161-9. doi: 10.1007/978-1-4939-6358-4_12. Methods Mol Biol. 2016. PMID: 27631805
-
Using Biotinylated SUMO-Traps to Analyze SUMOylated Proteins.Methods Mol Biol. 2016;1475:109-21. doi: 10.1007/978-1-4939-6358-4_8. Methods Mol Biol. 2016. PMID: 27631801
-
Role of ubiquitin-like proteins in transcriptional regulation.Ernst Schering Res Found Workshop. 2006;(57):173-92. doi: 10.1007/3-540-37633-x_10. Ernst Schering Res Found Workshop. 2006. PMID: 16568955 Review.
-
Sumoylation and other ubiquitin-like post-translational modifications in plants.Trends Cell Biol. 2010 Apr;20(4):223-32. doi: 10.1016/j.tcb.2010.01.007. Epub 2010 Feb 26. Trends Cell Biol. 2010. PMID: 20189809 Review.
-
Studying the ubiquitin code through biotin-based labelling methods.Semin Cell Dev Biol. 2022 Dec;132:109-119. doi: 10.1016/j.semcdb.2022.02.009. Epub 2022 Feb 16. Semin Cell Dev Biol. 2022. PMID: 35181195 Review.
Cited by
-
LUZP1, a novel regulator of primary cilia and the actin cytoskeleton, is a contributing factor in Townes-Brocks Syndrome.Elife. 2020 Jun 18;9:e55957. doi: 10.7554/eLife.55957. Elife. 2020. PMID: 32553112 Free PMC article.
-
Identification of proximal SUMO-dependent interactors using SUMO-ID.Nat Commun. 2021 Nov 18;12(1):6671. doi: 10.1038/s41467-021-26807-6. Nat Commun. 2021. PMID: 34795231 Free PMC article.
-
SUMO monoclonal antibodies vary in sensitivity, specificity, and ability to detect types of SUMO conjugate.Sci Rep. 2022 Dec 9;12(1):21343. doi: 10.1038/s41598-022-25665-6. Sci Rep. 2022. PMID: 36494414 Free PMC article.
-
P-Rex1 is a novel substrate of the E3 ubiquitin ligase Malin associated with Lafora disease.Neurobiol Dis. 2023 Feb;177:105998. doi: 10.1016/j.nbd.2023.105998. Epub 2023 Jan 10. Neurobiol Dis. 2023. PMID: 36638890 Free PMC article.
-
Novel insights into the interaction of UBA5 with UFM1 via a UFM1-interacting sequence.Sci Rep. 2017 Mar 30;7(1):508. doi: 10.1038/s41598-017-00610-0. Sci Rep. 2017. PMID: 28360427 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

