Mechanisms and regulation of DNA replication initiation in eukaryotes
- PMID: 28094588
- PMCID: PMC5545932
- DOI: 10.1080/10409238.2016.1274717
Mechanisms and regulation of DNA replication initiation in eukaryotes
Abstract
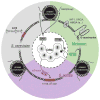
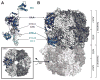
Cellular DNA replication is initiated through the action of multiprotein complexes that recognize replication start sites in the chromosome (termed origins) and facilitate duplex DNA melting within these regions. In a typical cell cycle, initiation occurs only once per origin and each round of replication is tightly coupled to cell division. To avoid aberrant origin firing and re-replication, eukaryotes tightly regulate two events in the initiation process: loading of the replicative helicase, MCM2-7, onto chromatin by the origin recognition complex (ORC), and subsequent activation of the helicase by its incorporation into a complex known as the CMG. Recent work has begun to reveal the details of an orchestrated and sequential exchange of initiation factors on DNA that give rise to a replication-competent complex, the replisome. Here, we review the molecular mechanisms that underpin eukaryotic DNA replication initiation - from selecting replication start sites to replicative helicase loading and activation - and describe how these events are often distinctly regulated across different eukaryotic model organisms.
Keywords: CMG; Cdc6; Cdt1; DNA replication; MCM2-7; ORC; helicase; initiator.
Figures

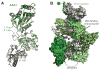

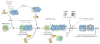

Similar articles
-
The dynamics of eukaryotic replication initiation: origin specificity, licensing, and firing at the single-molecule level.Mol Cell. 2015 May 7;58(3):483-94. doi: 10.1016/j.molcel.2015.03.017. Epub 2015 Apr 23. Mol Cell. 2015. PMID: 25921072 Free PMC article.
-
Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading.Cell. 2015 Apr 23;161(3):513-525. doi: 10.1016/j.cell.2015.03.012. Epub 2015 Apr 16. Cell. 2015. PMID: 25892223 Free PMC article.
-
Regulation of replication origin licensing by ORC phosphorylation reveals a two-step mechanism for Mcm2-7 ring closing.Proc Natl Acad Sci U S A. 2023 Jul 18;120(29):e2221484120. doi: 10.1073/pnas.2221484120. Epub 2023 Jul 10. Proc Natl Acad Sci U S A. 2023. PMID: 37428921 Free PMC article.
-
Eukaryotic DNA replication: Orchestrated action of multi-subunit protein complexes.Mutat Res. 2018 May;809:58-69. doi: 10.1016/j.mrfmmm.2017.04.002. Epub 2017 May 1. Mutat Res. 2018. PMID: 28501329 Review.
-
Unique Roles of the Non-identical MCM Subunits in DNA Replication Licensing.Mol Cell. 2017 Jul 20;67(2):168-179. doi: 10.1016/j.molcel.2017.06.016. Mol Cell. 2017. PMID: 28732205 Review.
Cited by
-
Recent advances in understanding DNA replication: cell type-specific adaptation of the DNA replication program.F1000Res. 2018 Aug 29;7:F1000 Faculty Rev-1351. doi: 10.12688/f1000research.15408.1. eCollection 2018. F1000Res. 2018. PMID: 30228862 Free PMC article. Review.
-
NFIB facilitates replication licensing by acting as a genome organizer.Nat Commun. 2023 Aug 21;14(1):5076. doi: 10.1038/s41467-023-40846-1. Nat Commun. 2023. PMID: 37604829 Free PMC article.
-
Chromatin and Nuclear Dynamics in the Maintenance of Replication Fork Integrity.Front Genet. 2021 Dec 14;12:773426. doi: 10.3389/fgene.2021.773426. eCollection 2021. Front Genet. 2021. PMID: 34970302 Free PMC article. Review.
-
An Orc1/Cdc6 ortholog functions as a key regulator in the DNA damage response in Archaea.Nucleic Acids Res. 2018 Jul 27;46(13):6697-6711. doi: 10.1093/nar/gky487. Nucleic Acids Res. 2018. PMID: 29878182 Free PMC article.
-
Mechanisms of hexameric helicases.Crit Rev Biochem Mol Biol. 2021 Dec;56(6):621-639. doi: 10.1080/10409238.2021.1954597. Epub 2021 Aug 17. Crit Rev Biochem Mol Biol. 2021. PMID: 34404299 Free PMC article. Review.
References
-
- Abreu CM, Kumar R, Hamilton D, Dawdy AW, Creavin K, Eivers S, Finn K, Balsbaugh JL, O’connor R, Kiely PA, Shabanowitz J, Hunt DF, Grenon M, Lowndes NF. Site-specific phosphorylation of the DNA damage response mediator rad9 by cyclin-dependent kinases regulates activation of checkpoint kinase 1. PLoS Genet. 2013;9:e1003310. - PMC - PubMed
-
- Adachi Y, Usukura J, Yanagida M. A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes Cells. 1997;2:467–79. - PubMed
-
- Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–6. - PubMed
-
- Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet. 2007;8:588–600. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
