sta-1 is repressed by mir-58 family in Caenorhabditis elegans
- PMID: 28090395
- PMCID: PMC5190142
- DOI: 10.1080/21624054.2016.1238560
sta-1 is repressed by mir-58 family in Caenorhabditis elegans
Abstract
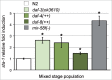
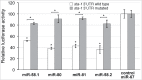
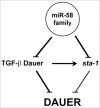
The miR-58 family comprises 6 microRNAs with largely shared functions, and with an overall high expression, because one of its members, miR-58, is the most abundant microRNA in Caenorhabditis elegans. We recently found that 2 TGF-β signaling pathways, Sma/Mab and Dauer, responsible for body size and dauer formation respectively, among other phenotypes, are downregulated by the miR-58 family. Here, we further explore this family by showing that it also acts through the sta-1 3'UTR. sta-1 encodes a transcription factor, homologous to mammalian STATs, that inhibits dauer formation in association with the TGF-β Dauer pathway. We also observe that mutants with a constitutively active TGF-β Dauer pathway express higher levels of sta-1 mRNA. Our results reinforce the view of the miR-58 family and STA-1 as regulators of dauer formation in coordination with the TGF-β Dauer pathway.
Keywords: Dauer; TGF-β Dauer; microRNAs; mir-58; sta-1.
Figures
Similar articles
-
miR-58 family and TGF-β pathways regulate each other in Caenorhabditis elegans.Nucleic Acids Res. 2015 Nov 16;43(20):9978-93. doi: 10.1093/nar/gkv923. Epub 2015 Sep 22. Nucleic Acids Res. 2015. PMID: 26400166 Free PMC article.
-
C. elegans STAT cooperates with DAF-7/TGF-beta signaling to repress dauer formation.Curr Biol. 2006 Jan 10;16(1):89-94. doi: 10.1016/j.cub.2005.11.061. Curr Biol. 2006. PMID: 16401427
-
Regulation of a hitchhiking behavior by neuronal insulin and TGF-β signaling in the nematode Caenorhabditis elegans.Biochem Biophys Res Commun. 2017 Mar 4;484(2):323-330. doi: 10.1016/j.bbrc.2017.01.113. Epub 2017 Jan 25. Biochem Biophys Res Commun. 2017. PMID: 28131836
-
lon-1 regulates Caenorhabditis elegans body size downstream of the dbl-1 TGF beta signaling pathway.Dev Biol. 2002 Jun 15;246(2):418-28. doi: 10.1006/dbio.2002.0662. Dev Biol. 2002. PMID: 12051826
-
Alternate metabolism during the dauer stage of the nematode Caenorhabditis elegans.Exp Gerontol. 2005 Nov;40(11):850-6. doi: 10.1016/j.exger.2005.09.006. Epub 2005 Oct 10. Exp Gerontol. 2005. PMID: 16221538 Review.
Cited by
-
The multifaceted roles of microRNAs in differentiation.Curr Opin Cell Biol. 2020 Dec;67:118-140. doi: 10.1016/j.ceb.2020.08.015. Epub 2020 Nov 3. Curr Opin Cell Biol. 2020. PMID: 33152557 Free PMC article. Review.
-
Current Status of Drug Targets and Emerging Therapeutic Strategies in the Management of Alzheimer's Disease.Curr Neuropharmacol. 2020;18(9):883-903. doi: 10.2174/1570159X18666200429011823. Curr Neuropharmacol. 2020. PMID: 32348223 Free PMC article. Review.
-
Recent Molecular Genetic Explorations of Caenorhabditis elegans MicroRNAs.Genetics. 2018 Jul;209(3):651-673. doi: 10.1534/genetics.118.300291. Genetics. 2018. PMID: 29967059 Free PMC article.
References
-
- Steinkraus BR, Toegel M, Fulga TA. Tiny giants of gene regulation: experimental strategies for microRNA functional studies. Wiley Interdiscip Rev Dev Biol 2016; 5:311-62; PMID:26950183; http://dx.doi.org/10.1002/wdev.223 - DOI - PMC - PubMed
-
- Jin Y, Chen Z, Liu X, Zhou X. Evaluating the microRNA targeting sites by luciferase reporter gene assay. Methods Mol Biol 2013; 936:117-27; PMID:23007504; http://dx.doi.org/10.1007/978-1-62703-083-0_10 - DOI - PMC - PubMed
-
- Alvarez ML. Faster experimental validation of microRNA targets using cold fusion cloning and a dual Firefly-Renilla luciferase reporter assay. Methods Mol Biol 2014; 1182:227-43; PMID:25055916; http://dx.doi.org/10.1007/978-1-4939-1062-5_21 - DOI - PubMed
-
- Chou C, Chang N, Shrestha S, Hsu S, Lin Y, Lee W, Yang C, Hong H, Wei T, Tu S, et al.. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 2016; 44:D239-47; PMID:26590260; http://dx.doi.org/10.1093/nar/gkv1258 - DOI - PMC - PubMed
-
- de Lucas MP, Sáez AG, Lozano E. miR-58 family and TGF-β pathways regulate each other in Caenorhabditis elegans. Nucleic Acids Res 2015; 43:9978-93; PMID:26400166; http://dx.doi.org/10.1093/nar/gkv923 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
