An Inducible Reconstitution System for the Real-Time Kinetic Analysis of Protease Activity and Inhibition Inside the Membrane
- PMID: 28065265
- PMCID: PMC5224909
- DOI: 10.1016/bs.mie.2016.10.025
An Inducible Reconstitution System for the Real-Time Kinetic Analysis of Protease Activity and Inhibition Inside the Membrane
Abstract
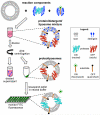
Intramembrane proteases are an ancient and diverse group of multispanning membrane proteins that cleave transmembrane substrates inside the membrane to effect a wide range of biological processes. As proteases, a clear understanding of their function requires kinetic dissection of their catalytic mechanism, but this is difficult to achieve for membrane proteins. Kinetic measurements in detergent systems are complicated by micelle fusion/exchange, which introduces an additional kinetic step and imposes system-specific behaviors (e.g., cooperativity). Conversely, kinetic analysis in proteoliposomes is hindered by premature substrate cleavage during coreconstitution, and lack of methods to quantify proteolysis in membranes in real time. In this chapter, we describe a method for the real-time kinetic analysis of intramembrane proteolysis in model liposomes. Our assay is inducible, because the enzyme is held inactive by low pH during reconstitution, and fluorogenic, since fluorescence emission from the substrate is quenched near lipids but restored upon proteolytic release from the membrane. The precise measurement of initial reaction velocities continuously in real time facilitates accurate steady-state kinetic analysis of intramembrane proteolysis and its inhibition inside the membrane environment. Using real data we describe a step-by-step strategy to implement this assay for essentially any intramembrane protease.
Keywords: Alzheimer's disease; Cancer; Cell signaling; ER-associated degradation; Gamma-secretase; Malaria; Membrane protein; Parkinson's disease; Presenilin; Protease; Regulated intramembrane proteolysis; Site-2 protease.
© 2017 Elsevier Inc. All rights reserved.
Figures

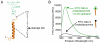
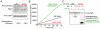
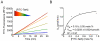

Similar articles
-
Understanding intramembrane proteolysis: from protein dynamics to reaction kinetics.Trends Biochem Sci. 2015 Jun;40(6):318-27. doi: 10.1016/j.tibs.2015.04.001. Epub 2015 May 1. Trends Biochem Sci. 2015. PMID: 25941170 Review.
-
Isolation of intramembrane proteases in membrane-like environments.Biochim Biophys Acta Biomembr. 2020 Apr 1;1862(4):183193. doi: 10.1016/j.bbamem.2020.183193. Epub 2020 Jan 13. Biochim Biophys Acta Biomembr. 2020. PMID: 31945321 Review.
-
Enzymatic Assays for Studying Intramembrane Proteolysis.Methods Enzymol. 2017;584:295-308. doi: 10.1016/bs.mie.2016.10.026. Epub 2016 Dec 24. Methods Enzymol. 2017. PMID: 28065268 Free PMC article.
-
Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity.Proc Natl Acad Sci U S A. 2005 Feb 8;102(6):1883-8. doi: 10.1073/pnas.0408306102. Epub 2005 Jan 31. Proc Natl Acad Sci U S A. 2005. PMID: 15684070 Free PMC article.
-
Expression, Purification, and Enzymatic Characterization of Intramembrane Proteases.Methods Enzymol. 2017;584:127-155. doi: 10.1016/bs.mie.2016.09.046. Epub 2016 Oct 27. Methods Enzymol. 2017. PMID: 28065261 Review.
Cited by
-
Rhomboid distorts lipids to break the viscosity-imposed speed limit of membrane diffusion.Science. 2019 Feb 1;363(6426):eaao0076. doi: 10.1126/science.aao0076. Science. 2019. PMID: 30705155 Free PMC article.
-
Single-Molecule Analyses Reveal Rhomboid Proteins Are Strict and Functional Monomers in the Membrane.Biophys J. 2018 Nov 6;115(9):1755-1761. doi: 10.1016/j.bpj.2018.09.024. Epub 2018 Oct 2. Biophys J. 2018. PMID: 30342748 Free PMC article.
-
Decoding the Functional Evolution of an Intramembrane Protease Superfamily by Statistical Coupling Analysis.Structure. 2020 Dec 1;28(12):1329-1336.e4. doi: 10.1016/j.str.2020.07.015. Epub 2020 Aug 13. Structure. 2020. PMID: 32795403 Free PMC article.
-
The opening dynamics of the lateral gate regulates the activity of rhomboid proteases.Sci Adv. 2023 Jul 21;9(29):eadh3858. doi: 10.1126/sciadv.adh3858. Epub 2023 Jul 19. Sci Adv. 2023. PMID: 37467320 Free PMC article.
-
Lipid-polymer nanoparticles to probe the native-like environment of intramembrane rhomboid protease GlpG and its activity.Nat Commun. 2024 Aug 30;15(1):7533. doi: 10.1038/s41467-024-51989-0. Nat Commun. 2024. PMID: 39215029 Free PMC article.
References
-
- Akiyama Y, Kanehara K, Ito K. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO Journal. 2004;23(22):4434–4442. http://dx.doi.org/10.1038/sj.emboj.7600449. - DOI - PMC - PubMed
-
- Baker RP, Urban S. Architectural and thermodynamic principles underlying intramembrane protease function. Nature Chemical Biology. 2012;8(9):759–768. http://dx.doi.org/10.1038/nchembio.1021. - DOI - PMC - PubMed
-
- Baker RP, Urban S. Cytosolic extensions directly regulate a rhomboid protease by modulating substrate gating. Nature. 2015;523(7558):101–105. http://dx.doi.org/10.1038/nature14357. - DOI - PMC - PubMed
-
- Bangham AD, Horne RW. Negative staining of phospholipids and their structural modification by surface-active agents as observed in the electron microscope. Journal of Molecular Biology. 1964;8(5):660–668. http://dx.doi.org/10.1016/S0022-2836(64)80115-7. - DOI - PubMed
-
- Barzykin AV, Tachiya M. Reaction kinetics in microdisperse systems with exchange. Journal of Physical Chemistry. 1994;98(10):2677–2687. http://dx.doi.org/10.1021/j100061a027. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

