Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast
- PMID: 28062837
- PMCID: PMC5555582
- DOI: 10.1042/BCJ20160516
Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast
Abstract
Ribosome biogenesis requires the intertwined processes of folding, modification, and processing of ribosomal RNA, together with binding of ribosomal proteins. In eukaryotic cells, ribosome assembly begins in the nucleolus, continues in the nucleoplasm, and is not completed until after nascent particles are exported to the cytoplasm. The efficiency and fidelity of ribosome biogenesis are facilitated by >200 assembly factors and ∼76 different small nucleolar RNAs. The pathway is driven forward by numerous remodeling events to rearrange the ribonucleoprotein architecture of pre-ribosomes. Here, we describe principles of ribosome assembly that have emerged from recent studies of biogenesis of the large ribosomal subunit in the yeast Saccharomyces cerevisiae We describe tools that have empowered investigations of ribosome biogenesis, and then summarize recent discoveries about each of the consecutive steps of subunit assembly.
Keywords: ribonucleoproteins; ribosome assembly; ribosomes; yeast.
© 2017 The Author(s); published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
The Authors declare that there are no competing interests associated with the manuscript.
Figures




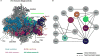
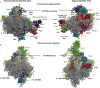
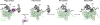
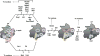
Similar articles
-
Assembly and early maturation of large subunit precursors.RNA. 2019 Apr;25(4):465-471. doi: 10.1261/rna.069799.118. Epub 2019 Jan 22. RNA. 2019. PMID: 30670483 Free PMC article.
-
Pol5 is required for recycling of small subunit biogenesis factors and for formation of the peptide exit tunnel of the large ribosomal subunit.Nucleic Acids Res. 2020 Jan 10;48(1):405-420. doi: 10.1093/nar/gkz1079. Nucleic Acids Res. 2020. PMID: 31745560 Free PMC article.
-
Role of the yeast ribosomal protein L16 in ribosome biogenesis.FEBS J. 2016 Aug;283(16):2968-85. doi: 10.1111/febs.13797. Epub 2016 Jul 15. FEBS J. 2016. PMID: 27374275
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
-
Ribosome assembly in eukaryotes.Gene. 2003 Aug 14;313:17-42. doi: 10.1016/s0378-1119(03)00629-2. Gene. 2003. PMID: 12957375 Review.
Cited by
-
Separation and Paired Proteome Profiling of Plant Chloroplast and Cytoplasmic Ribosomes.Plants (Basel). 2020 Jul 14;9(7):892. doi: 10.3390/plants9070892. Plants (Basel). 2020. PMID: 32674508 Free PMC article.
-
Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.Nat Struct Mol Biol. 2019 Sep;26(9):830-839. doi: 10.1038/s41594-019-0289-8. Epub 2019 Sep 5. Nat Struct Mol Biol. 2019. PMID: 31488907 Free PMC article.
-
Reverse Transcription-Quantitative Real-Time PCR (RT-qPCR) Assay for the Rapid Enumeration of Live Candida auris Cells from the Health Care Environment.J Clin Microbiol. 2022 Feb 16;60(2):e0077921. doi: 10.1128/JCM.00779-21. Epub 2021 Dec 8. J Clin Microbiol. 2022. PMID: 34878804 Free PMC article.
-
Inhibiting eukaryotic ribosome biogenesis.BMC Biol. 2019 Jun 10;17(1):46. doi: 10.1186/s12915-019-0664-2. BMC Biol. 2019. PMID: 31182083 Free PMC article.
-
Introns provide a platform for intergenic regulatory feedback of RPL22 paralogs in yeast.PLoS One. 2018 Jan 5;13(1):e0190685. doi: 10.1371/journal.pone.0190685. eCollection 2018. PLoS One. 2018. PMID: 29304067 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

