Structure(s), function(s), and inhibition of the RNA-dependent RNA polymerase of noroviruses
- PMID: 28041960
- PMCID: PMC7114559
- DOI: 10.1016/j.virusres.2016.12.018
Structure(s), function(s), and inhibition of the RNA-dependent RNA polymerase of noroviruses
Abstract
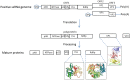
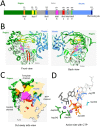
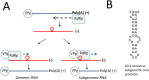
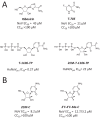
Noroviruses belong to the Caliciviridae family of single-stranded positive-sense RNA viruses. The genus Norovirus includes seven genogroups (designated GI-GVII), of which GI, GII and GIV infect humans. Human noroviruses are responsible for widespread outbreaks of acute gastroenteritis and represent one of the most common causes of foodborne illness. No vaccine or antiviral treatment options are available for norovirus infection. The RNA-dependent RNA polymerase (RdRp) of noroviruses is a key enzyme responsible for transcription and replication of the viral genome. Here, we review the progress made in understanding the structures and functions of norovirus RdRp and its use as a target for small molecule inhibitors. Crystal structures of the RdRp at different stages of substrate interaction have been determined, which shed light on its multi-step catalytic cycle. The in vitro assays and in vivo animal models that have been developed to identify and characterize inhibitors of norovirus RdRp are also summarized, followed by an update on the current antiviral research targeting different regions of norovirus RdRp. In the future, structure-based drug design and rational optimization of known nucleoside and non-nucleoside inhibitors of norovirus RdRp may pave the way towards the next generation of direct-acting antivirals.
Keywords: Caliciviridae; Non-nucleoside inhibitor; Norovirus; Nucleoside analogs; RNA-dependent RNA polymerase (RdRp); Virus inhibitors.
Copyright © 2016. Published by Elsevier B.V.
Figures

Similar articles
-
Naphthalene-sulfonate inhibitors of human norovirus RNA-dependent RNA-polymerase.Antiviral Res. 2014 Feb;102:23-8. doi: 10.1016/j.antiviral.2013.11.016. Epub 2013 Dec 4. Antiviral Res. 2014. PMID: 24316032
-
Broad-spectrum non-nucleoside inhibitors for caliciviruses.Antiviral Res. 2017 Oct;146:65-75. doi: 10.1016/j.antiviral.2017.07.014. Epub 2017 Jul 27. Antiviral Res. 2017. PMID: 28757394
-
Structure-based inhibition of Norovirus RNA-dependent RNA polymerases.J Mol Biol. 2012 Jun 8;419(3-4):198-210. doi: 10.1016/j.jmb.2012.03.008. Epub 2012 Mar 21. J Mol Biol. 2012. PMID: 22446684
-
Norovirus gastroenteritis, increased understanding and future antiviral options.Curr Opin Investig Drugs. 2008 Feb;9(2):146-51. Curr Opin Investig Drugs. 2008. PMID: 18246517 Review.
-
Recent Advances in the Discovery of Norovirus Therapeutics.J Med Chem. 2015 Dec 24;58(24):9438-50. doi: 10.1021/acs.jmedchem.5b00762. Epub 2015 Aug 17. J Med Chem. 2015. PMID: 26258852 Free PMC article. Review.
Cited by
-
Recent advances in the discovery of potent RNA-dependent RNA-polymerase (RdRp) inhibitors targeting viruses.RSC Med Chem. 2020 Dec 23;12(3):306-320. doi: 10.1039/d0md00318b. eCollection 2021 Mar 1. RSC Med Chem. 2020. PMID: 34046618 Free PMC article. Review.
-
Serological surveillance of GI norovirus reveals persistence of blockade antibody in a Jidong community-based prospective cohort, 2014-2018.Front Cell Infect Microbiol. 2023 Dec 18;13:1258550. doi: 10.3389/fcimb.2023.1258550. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38188632 Free PMC article.
-
Genomic analysis of human noroviruses using combined Illumina-Nanopore data.Virus Evol. 2021 Sep 15;7(2):veab079. doi: 10.1093/ve/veab079. eCollection 2021. Virus Evol. 2021. PMID: 35186325 Free PMC article.
-
Influence of traditional Chinese medicines on the in vivo metabolism of lopinavir/ritonavir based on UHPLC-MS/MS analysis.J Pharm Anal. 2022 Apr;12(2):270-277. doi: 10.1016/j.jpha.2021.06.006. Epub 2021 Jun 25. J Pharm Anal. 2022. PMID: 35582404 Free PMC article.
-
In silico identification of promising inhibitor against RNA-dependent RNA polymerase target of SARS-CoV-2.Mol Biol Res Commun. 2021 Sep;10(3):131-140. doi: 10.22099/mbrc.2021.40367.1621. Mol Biol Res Commun. 2021. PMID: 34476266 Free PMC article.
References
-
- Alam I., Lee J.H., Cho K.J., Han K.R., Yang J.M., Chung M.S., Kim K.H. Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase in complex with 2-thiouridine or ribavirin. Virology. 2012;426(2):143–151. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

