Severe Symptomatic Primary Human Cytomegalovirus Infection despite Effective Innate and Adaptive Immune Responses
- PMID: 28031361
- PMCID: PMC5309965
- DOI: 10.1128/JVI.02245-16
Severe Symptomatic Primary Human Cytomegalovirus Infection despite Effective Innate and Adaptive Immune Responses
Abstract
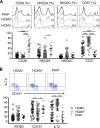
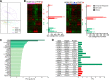
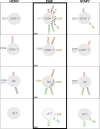
Primary human cytomegalovirus (HCMV) infection usually goes unnoticed, causing mild or no symptoms in immunocompetent individuals. However, some rare severe clinical cases have been reported without investigation of host immune responses or viral virulence. In the present study, we investigate for the first time phenotypic and functional features, together with gene expression profiles in immunocompetent adults experiencing a severe primary HCMV infection. Twenty primary HCMV-infected patients (PHIP) were enrolled, as well as 26 HCMV-seronegative and 39 HCMV-seropositive healthy controls. PHIP had extensive lymphocytosis marked by massive expansion of natural killer (NK) and T cell compartments. Interestingly, PHIP mounted efficient innate and adaptive immune responses with a deep HCMV imprint, revealed mainly by the expansion of NKG2C+ NK cells, CD16+ Vδ2(-) γδ T cells, and conventional HCMV-specific CD8+ T cells. The main effector lymphocytes were activated and displayed an early immune phenotype that developed toward a more mature differentiated status. We suggest that both massive lymphocytosis and excessive lymphocyte activation could contribute to massive cytokine production, known to mediate tissue damage observed in PHIP. Taken together, these findings bring new insights into the comprehensive understanding of immune mechanisms involved during primary HCMV infection in immunocompetent individuals.IMPORTANCE HCMV-specific immune responses have been extensively documented in immunocompromised patients and during in utero acquisition. While it usually goes unnoticed, some rare severe clinical cases of primary HCMV infection have been reported in immunocompetent patients. However, host immune responses or HCMV virulence in these patients has not so far been investigated. In the present study, we show massive expansion of NK and T cell compartments during the symptomatic stage of acute HCMV infection. The patients mounted efficient innate and adaptive immune responses with a deep HCMV imprint. The massive lymphocytosis could be the result of nonadapted or uncontrolled immune responses limiting the effectiveness of the specific responses mounted. Both massive lymphocytosis and excessive lymphocyte activation could contribute to massive cytokine production, known to mediate tissue damage. Furthermore, we cannot exclude a delayed immune response caused by immune escape established by HCMV strains.
Keywords: HCMV; NK cells; T lymphocytes; adaptive immunity; innate immunity.
Copyright © 2017 American Society for Microbiology.
Figures




Similar articles
-
NKG2C(+)CD57(+) Natural Killer Cell Expansion Parallels Cytomegalovirus-Specific CD8(+) T Cell Evolution towards Senescence.J Immunol Res. 2016;2016:7470124. doi: 10.1155/2016/7470124. Epub 2016 May 29. J Immunol Res. 2016. PMID: 27314055 Free PMC article.
-
Human Cytomegalovirus Particles Treated with Specific Antibodies Induce Intrinsic and Adaptive but Not Innate Immune Responses.J Virol. 2017 Oct 27;91(22):e00678-17. doi: 10.1128/JVI.00678-17. Print 2017 Nov 15. J Virol. 2017. PMID: 28878085 Free PMC article.
-
Late Development of FcεRγneg Adaptive Natural Killer Cells Upon Human Cytomegalovirus Reactivation in Umbilical Cord Blood Transplantation Recipients.Front Immunol. 2018 May 15;9:1050. doi: 10.3389/fimmu.2018.01050. eCollection 2018. Front Immunol. 2018. PMID: 29868012 Free PMC article.
-
The CD94/NKG2C+ NK-cell subset on the edge of innate and adaptive immunity to human cytomegalovirus infection.Semin Immunol. 2014 Apr;26(2):145-51. doi: 10.1016/j.smim.2014.03.002. Epub 2014 Mar 22. Semin Immunol. 2014. PMID: 24666761 Review.
-
Molecular characterization of HCMV-specific immune responses: Parallels between CD8(+) T cells, CD4(+) T cells, and NK cells.Eur J Immunol. 2015 Sep;45(9):2433-45. doi: 10.1002/eji.201545495. Epub 2015 Aug 24. Eur J Immunol. 2015. PMID: 26228786 Review.
Cited by
-
The CD4+ T Cell Response to Human Cytomegalovirus in Healthy and Immunocompromised People.Front Cell Infect Microbiol. 2020 May 19;10:202. doi: 10.3389/fcimb.2020.00202. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32509591 Free PMC article. Review.
-
Advances in the treatment of cytomegalovirus.Br Med Bull. 2019 Sep 19;131(1):5-17. doi: 10.1093/bmb/ldz031. Br Med Bull. 2019. PMID: 31580403 Free PMC article.
-
Cell type-specific biogenesis of novel vesicles containing viral products in human cytomegalovirus infection.J Virol. 2021 May 10;95(11):e02358-20. doi: 10.1128/JVI.02358-20. Epub 2021 Mar 24. J Virol. 2021. PMID: 33762413 Free PMC article.
-
Shared Molecular Signatures Across Neurodegenerative Diseases and Herpes Virus Infections Highlights Potential Mechanisms for Maladaptive Innate Immune Responses.Sci Rep. 2019 Jun 19;9(1):8795. doi: 10.1038/s41598-019-45129-8. Sci Rep. 2019. PMID: 31217489 Free PMC article.
-
Immunotherapy with adoptive cytomegalovirus-specific T cells transfer: Summarizing latest gene engineering techniques.Health Sci Rep. 2021 Jul 8;4(3):e322. doi: 10.1002/hsr2.322. eCollection 2021 Sep. Health Sci Rep. 2021. PMID: 34263085 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

