Reverse Genetics Approaches for the Development of Influenza Vaccines
- PMID: 28025504
- PMCID: PMC5297655
- DOI: 10.3390/ijms18010020
Reverse Genetics Approaches for the Development of Influenza Vaccines
Abstract
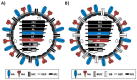
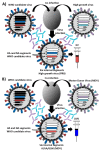
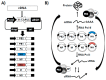
Influenza viruses cause annual seasonal epidemics and occasional pandemics of human respiratory disease. Influenza virus infections represent a serious public health and economic problem, which are most effectively prevented through vaccination. However, influenza viruses undergo continual antigenic variation, which requires either the annual reformulation of seasonal influenza vaccines or the rapid generation of vaccines against potential pandemic virus strains. The segmented nature of influenza virus allows for the reassortment between two or more viruses within a co-infected cell, and this characteristic has also been harnessed in the laboratory to generate reassortant viruses for their use as either inactivated or live-attenuated influenza vaccines. With the implementation of plasmid-based reverse genetics techniques, it is now possible to engineer recombinant influenza viruses entirely from full-length complementary DNA copies of the viral genome by transfection of susceptible cells. These reverse genetics systems have provided investigators with novel and powerful approaches to answer important questions about the biology of influenza viruses, including the function of viral proteins, their interaction with cellular host factors and the mechanisms of influenza virus transmission and pathogenesis. In addition, reverse genetics techniques have allowed the generation of recombinant influenza viruses, providing a powerful technology to develop both inactivated and live-attenuated influenza vaccines. In this review, we will summarize the current knowledge of state-of-the-art, plasmid-based, influenza reverse genetics approaches and their implementation to provide rapid, convenient, safe and more effective influenza inactivated or live-attenuated vaccines.
Keywords: antivirals; influenza inactivated virus; influenza vaccines; influenza virus; live-attenuated influenza virus; recombinant influenza virus; reverse genetics; universal vaccines; vaccination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Novel Approaches for The Development of Live Attenuated Influenza Vaccines.Viruses. 2019 Feb 22;11(2):190. doi: 10.3390/v11020190. Viruses. 2019. PMID: 30813325 Free PMC article. Review.
-
Influenza vaccines generated by reverse genetics.Curr Top Microbiol Immunol. 2004;283:313-42. doi: 10.1007/978-3-662-06099-5_9. Curr Top Microbiol Immunol. 2004. PMID: 15298174 Review.
-
Reverse Genetics of Influenza B Viruses.Methods Mol Biol. 2017;1602:205-238. doi: 10.1007/978-1-4939-6964-7_14. Methods Mol Biol. 2017. PMID: 28508223
-
[Using reverse genetics method for developing recombinant strains of influenza viruses acceptable for use as live attenuated vaccines].Zh Mikrobiol Epidemiol Immunobiol. 2009 Mar-Apr;(2):111-7. Zh Mikrobiol Epidemiol Immunobiol. 2009. PMID: 19462520 Review. Russian.
-
Reverse genetics for influenza B viruses and recent advances in vaccine development.Curr Opin Virol. 2020 Oct;44:191-202. doi: 10.1016/j.coviro.2020.10.005. Epub 2020 Nov 27. Curr Opin Virol. 2020. PMID: 33254031 Free PMC article. Review.
Cited by
-
Alternative Experimental Models for Studying Influenza Proteins, Host-Virus Interactions and Anti-Influenza Drugs.Pharmaceuticals (Basel). 2019 Sep 30;12(4):147. doi: 10.3390/ph12040147. Pharmaceuticals (Basel). 2019. PMID: 31575020 Free PMC article. Review.
-
Functional Characterization and Direct Comparison of Influenza A, B, C, and D NS1 Proteins in vitro and in vivo.Front Microbiol. 2019 Dec 17;10:2862. doi: 10.3389/fmicb.2019.02862. eCollection 2019. Front Microbiol. 2019. PMID: 31921042 Free PMC article.
-
Decoding antimicrobial resistance: unraveling molecular mechanisms and targeted strategies.Arch Microbiol. 2024 May 28;206(6):280. doi: 10.1007/s00203-024-03998-2. Arch Microbiol. 2024. PMID: 38805035 Review.
-
Mutation L319Q in the PB1 Polymerase Subunit Improves Attenuation of a Candidate Live-Attenuated Influenza A Virus Vaccine.Microbiol Spectr. 2022 Jun 29;10(3):e0007822. doi: 10.1128/spectrum.00078-22. Epub 2022 May 18. Microbiol Spectr. 2022. PMID: 35583364 Free PMC article.
-
Interplay of PA-X and NS1 Proteins in Replication and Pathogenesis of a Temperature-Sensitive 2009 Pandemic H1N1 Influenza A Virus.J Virol. 2017 Aug 10;91(17):e00720-17. doi: 10.1128/JVI.00720-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28637750 Free PMC article.
References
-
- Palese P.S., Shaw M.L. Orthomyxoviridae: The viruses and their replication. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., editors. Fields Virology. 5th ed. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2007.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

