Regulation of ULK1 Expression and Autophagy by STAT1
- PMID: 28011640
- PMCID: PMC5290961
- DOI: 10.1074/jbc.M116.771584
Regulation of ULK1 Expression and Autophagy by STAT1
Abstract
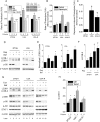
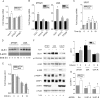
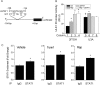
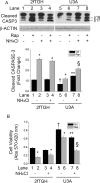
Autophagy involves the lysosomal degradation of cytoplasmic contents for regeneration of anabolic substrates during nutritional or inflammatory stress. Its initiation occurs rapidly after inactivation of the protein kinase mammalian target of rapamycin (mTOR) (or mechanistic target of rapamycin), leading to dephosphorylation of Unc-51-like kinase 1 (ULK1) and autophagosome formation. Recent studies indicate that mTOR can, in parallel, regulate the activity of stress transcription factors, including signal transducer and activator of transcription-1 (STAT1). The current study addresses the role of STAT1 as a transcriptional suppressor of autophagy genes and autophagic activity. We show that STAT1-deficient human fibrosarcoma cells exhibited enhanced autophagic flux as well as its induction by pharmacological inhibition of mTOR. Consistent with enhanced autophagy initiation, ULK1 mRNA and protein levels were increased in STAT1-deficient cells. By chromatin immunoprecipitation, STAT1 bound a putative regulatory sequence in the ULK1 5'-flanking region, the mutation of which increased ULK1 promoter activity, and rendered it unresponsive to mTOR inhibition. Consistent with an anti-apoptotic effect of autophagy, rapamycin-induced apoptosis and cytotoxicity were blocked in STAT1-deficient cells but restored in cells simultaneously exposed to the autophagy inhibitor ammonium chloride. In vivo, skeletal muscle ULK1 mRNA and protein levels as well as autophagic flux were significantly enhanced in STAT1-deficient mice. These results demonstrate a novel mechanism by which STAT1 negatively regulates ULK1 expression and autophagy.
Keywords: apoptosis; autophagy; mammalian target of rapamycin (mTOR); proteolysis; signal transducers and activators of transcription 1 (STAT1).
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
Similar articles
-
α-Synuclein disrupts microglial autophagy through STAT1-dependent suppression of Ulk1 transcription.J Neuroinflammation. 2024 Oct 26;21(1):275. doi: 10.1186/s12974-024-03268-4. J Neuroinflammation. 2024. PMID: 39462396 Free PMC article.
-
Mir214-3p and Hnf4a/Hnf4α reciprocally regulate Ulk1 expression and autophagy in nonalcoholic hepatic steatosis.Autophagy. 2021 Sep;17(9):2415-2431. doi: 10.1080/15548627.2020.1827779. Epub 2020 Oct 20. Autophagy. 2021. PMID: 33078654 Free PMC article.
-
Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis.Ann Rheum Dis. 2015 Jul;74(7):1432-40. doi: 10.1136/annrheumdis-2013-204599. Epub 2014 Mar 20. Ann Rheum Dis. 2015. PMID: 24651621
-
Function and regulation of ULK1: From physiology to pathology.Gene. 2022 Oct 5;840:146772. doi: 10.1016/j.gene.2022.146772. Epub 2022 Jul 26. Gene. 2022. PMID: 35905845 Review.
-
The mammalian ULK1 complex and autophagy initiation.Essays Biochem. 2017 Dec 12;61(6):585-596. doi: 10.1042/EBC20170021. Print 2017 Dec 12. Essays Biochem. 2017. PMID: 29233870 Free PMC article. Review.
Cited by
-
Inhibition of autophagosome-lysosome fusion contributes to TDCIPP-induced Aβ1-42 production in N2a-APPswe cells.Heliyon. 2024 Feb 21;10(8):e26832. doi: 10.1016/j.heliyon.2024.e26832. eCollection 2024 Apr 30. Heliyon. 2024. PMID: 38628727 Free PMC article.
-
Machine-Learning-Based Identification of Key Feature RNA-Signature Linked to Diagnosis of Hepatocellular Carcinoma.J Clin Exp Hepatol. 2024 Nov-Dec;14(6):101456. doi: 10.1016/j.jceh.2024.101456. Epub 2024 Jun 14. J Clin Exp Hepatol. 2024. PMID: 39055616
-
Autophagy Promotes Enrichment of Raft Components within Extracellular Vesicles Secreted by Human 2FTGH Cells.Int J Mol Sci. 2024 Jun 4;25(11):6175. doi: 10.3390/ijms25116175. Int J Mol Sci. 2024. PMID: 38892363 Free PMC article.
-
α-Synuclein disrupts microglial autophagy through STAT1-dependent suppression of Ulk1 transcription.J Neuroinflammation. 2024 Oct 26;21(1):275. doi: 10.1186/s12974-024-03268-4. J Neuroinflammation. 2024. PMID: 39462396 Free PMC article.
-
Molecular Regulation of Skeletal Muscle Growth and Organelle Biosynthesis: Practical Recommendations for Exercise Training.Int J Mol Sci. 2021 Mar 8;22(5):2741. doi: 10.3390/ijms22052741. Int J Mol Sci. 2021. PMID: 33800501 Free PMC article. Review.
References
-
- Mizushima N., and Komatsu M. (2011) Autophagy: renovation of cells and tissues. Cell 147, 728–741 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

