ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons
- PMID: 28008922
- PMCID: PMC5196439
- DOI: 10.1038/ncomms14021
ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons
Abstract
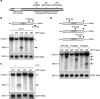
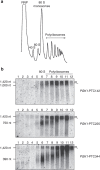
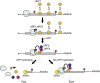
Nonsense-mediated mRNA decay (NMD) represents a eukaryotic quality control pathway that recognizes and rapidly degrades transcripts harbouring nonsense mutations to limit accumulation of non-functional and potentially toxic truncated polypeptides. A critical component of the NMD machinery is UPF1, an RNA helicase whose ATPase activity is essential for NMD, but for which the precise function and site of action remain unclear. We provide evidence that ATP hydrolysis by UPF1 is required for efficient translation termination and ribosome release at a premature termination codon. UPF1 ATPase mutants accumulate 3' RNA decay fragments harbouring a ribosome stalled during premature termination that impedes complete degradation of the mRNA. The ability of UPF1 to impinge on premature termination, moreover, requires ATP-binding, RNA-binding and NMD cofactors UPF2 and UPF3. Our results reveal that ATP hydrolysis by UPF1 modulates a functional interaction between the NMD machinery and terminating ribosomes necessary for targeting substrates to accelerated degradation.
Figures


Similar articles
-
Inhibition of post-termination ribosome recycling at premature termination codons in UPF1 ATPase mutants.Elife. 2020 Jul 22;9:e57834. doi: 10.7554/eLife.57834. Elife. 2020. PMID: 32697194 Free PMC article.
-
Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae.RNA. 2004 Apr;10(4):691-703. doi: 10.1261/rna.5147804. RNA. 2004. PMID: 15037778 Free PMC article.
-
Inactivation of NMD increases viability of sup45 nonsense mutants in Saccharomyces cerevisiae.BMC Mol Biol. 2007 Aug 16;8:71. doi: 10.1186/1471-2199-8-71. BMC Mol Biol. 2007. PMID: 17705828 Free PMC article.
-
Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay.Mol Biol Rep. 2018 Feb;45(1):39-55. doi: 10.1007/s11033-017-4139-7. Epub 2017 Dec 27. Mol Biol Rep. 2018. PMID: 29282598 Review.
-
Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay.Genes Cells. 2013 Mar;18(3):161-75. doi: 10.1111/gtc.12033. Epub 2013 Jan 28. Genes Cells. 2013. PMID: 23356578 Review.
Cited by
-
UPF1 mutants with intact ATPase but deficient helicase activities promote efficient nonsense-mediated mRNA decay.Nucleic Acids Res. 2022 Nov 11;50(20):11876-11894. doi: 10.1093/nar/gkac1026. Nucleic Acids Res. 2022. PMID: 36370101 Free PMC article.
-
The ribosomal stalk protein is crucial for the action of the conserved ATPase ABCE1.Nucleic Acids Res. 2018 Sep 6;46(15):7820-7830. doi: 10.1093/nar/gky619. Nucleic Acids Res. 2018. PMID: 30010948 Free PMC article.
-
Nonsense-mediated mRNA decay factors cure most [PSI+] prion variants.Proc Natl Acad Sci U S A. 2018 Feb 6;115(6):E1184-E1193. doi: 10.1073/pnas.1717495115. Epub 2018 Jan 22. Proc Natl Acad Sci U S A. 2018. PMID: 29358398 Free PMC article.
-
Translational suppression via IFG-1/eIF4G inhibits stress-induced RNA alternative splicing in Caenorhabditis elegans.Genetics. 2022 Jul 4;221(3):iyac075. doi: 10.1093/genetics/iyac075. Genetics. 2022. PMID: 35536193 Free PMC article.
-
Nonsense-Mediated mRNA Decay in Human Health and Diseases: Current Understanding, Regulatory Mechanisms and Future Perspectives.Mol Biotechnol. 2024 Sep 12. doi: 10.1007/s12033-024-01267-7. Online ahead of print. Mol Biotechnol. 2024. PMID: 39264527 Review.
References
-
- Lykke-Andersen S. & Jensen T. H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell. Biol. 15, 665–677 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

