General Amino Acid Control and 14-3-3 Proteins Bmh1/2 Are Required for Nitrogen Catabolite Repression-Sensitive Regulation of Gln3 and Gat1 Localization
- PMID: 28007891
- PMCID: PMC5289842
- DOI: 10.1534/genetics.116.195800
General Amino Acid Control and 14-3-3 Proteins Bmh1/2 Are Required for Nitrogen Catabolite Repression-Sensitive Regulation of Gln3 and Gat1 Localization
Abstract
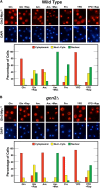
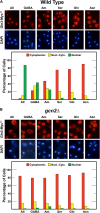
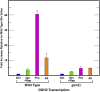
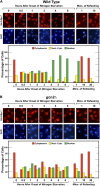
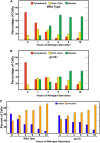
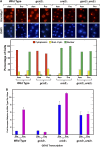
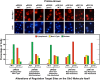
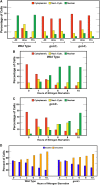
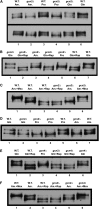
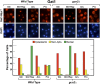
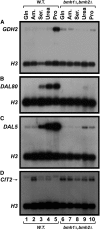
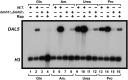
Nitrogen catabolite repression (NCR), the ability of Saccharomyces cerevisiae to use good nitrogen sources in preference to poor ones, derives from nitrogen-responsive regulation of the GATA family transcription activators Gln3 and Gat1 In nitrogen-replete conditions, the GATA factors are cytoplasmic and NCR-sensitive transcription minimal. When only poor nitrogen sources are available, Gln3 is nuclear, dramatically increasing GATA factor-mediated transcription. This regulation was originally attributed to mechanistic Tor protein kinase complex 1 (mTorC1)-mediated control of Gln3 However, we recently showed that two regulatory systems act cumulatively to maintain cytoplasmic Gln3 sequestration, only one of which is mTorC1. Present experiments demonstrate that the other previously elusive component is uncharged transfer RNA-activated, Gcn2 protein kinase-mediated general amino acid control (GAAC). Gcn2 and Gcn4 are required for NCR-sensitive nuclear Gln3-Myc13 localization, and from epistasis experiments Gcn2 appears to function upstream of Ure2 Bmh1/2 are also required for nuclear Gln3-Myc13 localization and appear to function downstream of Ure2 Overall, Gln3 phosphorylation levels decrease upon loss of Gcn2, Gcn4, or Bmh1/2 Our results add a new dimension to nitrogen-responsive GATA-factor regulation and demonstrate the cumulative participation of the mTorC1 and GAAC pathways, which respond oppositely to nitrogen availability, in the nitrogen-responsive control of catabolic gene expression in yeast.
Keywords: Bmh1/2; Gat1; Gcn2; Gln3; nitrogen catabolite repression.
Copyright © 2017 by the Genetics Society of America.
Figures
Similar articles
-
More than One Way in: Three Gln3 Sequences Required To Relieve Negative Ure2 Regulation and Support Nuclear Gln3 Import in Saccharomyces cerevisiae.Genetics. 2018 Jan;208(1):207-227. doi: 10.1534/genetics.117.300457. Epub 2017 Nov 7. Genetics. 2018. PMID: 29113979 Free PMC article.
-
Tor pathway control of the nitrogen-responsive DAL5 gene bifurcates at the level of Gln3 and Gat1 regulation in Saccharomyces cerevisiae.J Biol Chem. 2008 Apr 4;283(14):8919-29. doi: 10.1074/jbc.M708811200. Epub 2008 Feb 1. J Biol Chem. 2008. PMID: 18245087 Free PMC article.
-
GATA Factor Regulation in Excess Nitrogen Occurs Independently of Gtr-Ego Complex-Dependent TorC1 Activation.G3 (Bethesda). 2015 May 29;5(8):1625-38. doi: 10.1534/g3.115.019307. G3 (Bethesda). 2015. PMID: 26024867 Free PMC article.
-
Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots.FEMS Microbiol Rev. 2002 Aug;26(3):223-38. doi: 10.1111/j.1574-6976.2002.tb00612.x. FEMS Microbiol Rev. 2002. PMID: 12165425 Free PMC article. Review.
-
New functions of protein kinase Gcn2 in yeast and mammals.IUBMB Life. 2012 Dec;64(12):971-4. doi: 10.1002/iub.1090. Epub 2012 Nov 5. IUBMB Life. 2012. PMID: 23129244 Review.
Cited by
-
More than One Way in: Three Gln3 Sequences Required To Relieve Negative Ure2 Regulation and Support Nuclear Gln3 Import in Saccharomyces cerevisiae.Genetics. 2018 Jan;208(1):207-227. doi: 10.1534/genetics.117.300457. Epub 2017 Nov 7. Genetics. 2018. PMID: 29113979 Free PMC article.
-
The yeast 14-3-3 proteins Bmh1 and Bmh2 regulate key signaling pathways.Front Mol Biosci. 2024 Jan 24;11:1327014. doi: 10.3389/fmolb.2024.1327014. eCollection 2024. Front Mol Biosci. 2024. PMID: 38328397 Free PMC article. Review.
-
The flavonoid 4,4'-dimethoxychalcone promotes autophagy-dependent longevity across species.Nat Commun. 2019 Feb 19;10(1):651. doi: 10.1038/s41467-019-08555-w. Nat Commun. 2019. PMID: 30783116 Free PMC article.
-
A Gene from Ganoderma lucidum with Similarity to nmrA of Filamentous Ascomycetes Contributes to Regulating AreA.J Fungi (Basel). 2023 Apr 26;9(5):516. doi: 10.3390/jof9050516. J Fungi (Basel). 2023. PMID: 37233227 Free PMC article.
-
N- and C-terminal Gln3-Tor1 interaction sites: one acting negatively and the other positively to regulate nuclear Gln3 localization.Genetics. 2021 Apr 15;217(4):iyab017. doi: 10.1093/genetics/iyab017. Genetics. 2021. PMID: 33857304 Free PMC article.
References
-
- Beck T., Hall M. N., 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692. - PubMed
-
- Bertram P. G., Choi J. H., Carvalho J., Ai W., Zeng C., et al. , 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275: 35727–35733. - PubMed
-
- Binda M., Péli-Gulli M. P., Bonfils G., Panchaud N., Urban J., et al. , 2009. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35: 563–573. - PubMed
-
- Binda M., Bonfils G., Panchaud N., Péli-Gulli M. P., De Virgilio C., 2010. An EGOcentric view of TORC1 signaling. Cell Cycle 9: 221–222. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

