Human Cytomegalovirus Requires Epidermal Growth Factor Receptor Signaling To Enter and Initiate the Early Steps in the Establishment of Latency in CD34+ Human Progenitor Cells
- PMID: 27974567
- PMCID: PMC5309964
- DOI: 10.1128/JVI.01206-16
Human Cytomegalovirus Requires Epidermal Growth Factor Receptor Signaling To Enter and Initiate the Early Steps in the Establishment of Latency in CD34+ Human Progenitor Cells
Abstract
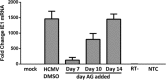
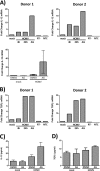
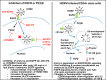
The establishment of human cytomegalovirus (HCMV) latency and persistence relies on the successful infection of hematopoietic cells, which serve as sites of viral persistence and contribute to viral spread. Here, using blocking antibodies and pharmacological inhibitors, we document that HCMV activation of the epidermal growth factor receptor (EGFR) and downstream phosphatidylinositol 3-kinase (PI3K) mediates viral entry into CD34+ human progenitor cells (HPCs), resulting in distinct cellular trafficking and nuclear translocation of the virus compared to that in other immune cells, such as we have documented in monocytes. We argue that the EGFR allows HCMV to regulate the cellular functions of these replication-restricted cells via its signaling activity following viral binding. In addition to regulating HCMV entry/trafficking, EGFR signaling may also shape the early steps required for the successful establishment of viral latency in CD34+ cells, as pharmacological inhibition of EGFR increases the transcription of lytic IE1/IE2 mRNA while curbing the expression of latency-associated UL138 mRNA. EGFR signaling following infection of CD34+ HPCs may also contribute to changes in hematopoietic potential, as treatment with the EGFR kinase (EGFRK) inhibitor AG1478 alters the expression of the cellular hematopoietic cytokine interleukin 12 (IL-12) in HCMV-infected cells but not in mock-infected cells. These findings, along with our previous work with monocytes, suggest that EGFR likely serves as an important determinant of HCMV tropism for select subsets of hematopoietic cells. Moreover, our new data suggest that EGFR is a key receptor for efficient viral entry and that the ensuing signaling regulates important early events required for successful infection of CD34+ HPCs by HCMV.IMPORTANCE HCMV establishes lifelong persistence within the majority of the human population without causing overt pathogenesis in healthy individuals. Despite this, reactivation of HCMV from its latent reservoir in the bone marrow causes significant morbidity and mortality in immunologically compromised individuals, such as bone marrow and solid organ transplant patients. Lifelong persistent infection has also been linked with the development of various cardiovascular diseases in otherwise healthy individuals. Current HCMV therapeutics target lytic replication, but not the latent viral reservoir; thus, an understanding of the molecular basis for viral latency and persistence is paramount to controlling or eliminating HCMV infection. Here, we show that the viral signalosome activated by HCMV binding to its entry receptor, EGFR, in CD34+ HPCs initiates early events necessary for successful latent infection of this cell type. EGFR and associated signaling players may therefore represent promising targets for mitigating HCMV persistence.
Keywords: CD34+ HPC; EGFR; HCMV; latency; virus entry.
Copyright © 2017 American Society for Microbiology.
Figures
Similar articles
-
Human Cytomegalovirus US28 Ligand Binding Activity Is Required for Latency in CD34+ Hematopoietic Progenitor Cells and Humanized NSG Mice.mBio. 2019 Aug 20;10(4):e01889-19. doi: 10.1128/mBio.01889-19. mBio. 2019. PMID: 31431555 Free PMC article.
-
CD34+ Hematopoietic Progenitor Cell Subsets Exhibit Differential Ability To Maintain Human Cytomegalovirus Latency and Persistence.J Virol. 2021 Jan 13;95(3):e02105-20. doi: 10.1128/JVI.02105-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33177198 Free PMC article.
-
Human Cytomegalovirus UL135 Interacts with Host Adaptor Proteins To Regulate Epidermal Growth Factor Receptor and Reactivation from Latency.J Virol. 2018 Sep 26;92(20):e00919-18. doi: 10.1128/JVI.00919-18. Print 2018 Oct 15. J Virol. 2018. PMID: 30089695 Free PMC article.
-
Cell signaling and cytomegalovirus reactivation: what do Src family kinases have to do with it?Biochem Soc Trans. 2020 Apr 29;48(2):667-675. doi: 10.1042/BST20191110. Biochem Soc Trans. 2020. PMID: 32311019 Free PMC article. Review.
-
Human Cytomegalovirus Host Interactions: EGFR and Host Cell Signaling Is a Point of Convergence Between Viral Infection and Functional Changes in Infected Cells.Front Microbiol. 2021 May 7;12:660901. doi: 10.3389/fmicb.2021.660901. eCollection 2021. Front Microbiol. 2021. PMID: 34025614 Free PMC article. Review.
Cited by
-
The Pentamer glycoprotein complex inhibits viral Immediate Early transcription during Human Cytomegalovirus infections.Proc Natl Acad Sci U S A. 2024 Sep 24;121(39):e2408078121. doi: 10.1073/pnas.2408078121. Epub 2024 Sep 18. Proc Natl Acad Sci U S A. 2024. PMID: 39292744
-
Breast milk induces the differentiation of monocytes into macrophages, promoting human cytomegalovirus infection.J Virol. 2024 Sep 17;98(9):e0117724. doi: 10.1128/jvi.01177-24. Epub 2024 Aug 28. J Virol. 2024. PMID: 39194236
-
Delivery of US28 by incoming HCMV particles rapidly attenuates Akt activity to suppress HCMV lytic replication in monocytes.Sci Signal. 2024 Aug 27;17(851):eadn8727. doi: 10.1126/scisignal.adn8727. Epub 2024 Aug 27. Sci Signal. 2024. PMID: 39190708 Free PMC article.
-
Human cytomegalovirus and neonatal infection.Curr Res Microb Sci. 2024 Jun 24;7:100257. doi: 10.1016/j.crmicr.2024.100257. eCollection 2024. Curr Res Microb Sci. 2024. PMID: 39070527 Free PMC article. Review.
-
Viral and host network analysis of the human cytomegalovirus transcriptome in latency.bioRxiv [Preprint]. 2024 May 21:2024.05.21.594597. doi: 10.1101/2024.05.21.594597. bioRxiv. 2024. PMID: 38826434 Free PMC article. Preprint.
References
-
- Mocarski E Jr, Shenk T, Griffiths PD, Pass R. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA, USA.
-
- Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol 325:417–470. - PubMed
-
- Nogalski MT, Collins-McMillen D, Yurochko AD. 2014. Overview of human cytomegalovirus pathogenesis, p 15–28. In Yurochko AD, Miller WE (ed), Human cytomegaloviruses: methods and protocols. Humana Press, New York, NY. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

