Tissue memory B cell repertoire analysis after ALVAC/AIDSVAX B/E gp120 immunization of rhesus macaques
- PMID: 27942585
- PMCID: PMC5135278
- DOI: 10.1172/jci.insight.88522
Tissue memory B cell repertoire analysis after ALVAC/AIDSVAX B/E gp120 immunization of rhesus macaques
Abstract
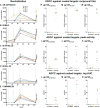
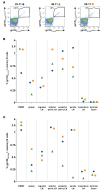
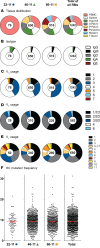
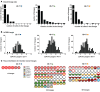
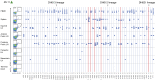
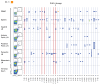
The ALVAC prime/ALVAC + AIDSVAX B/E boost RV144 vaccine trial induced an estimated 31% efficacy in a low-risk cohort where HIV‑1 exposures were likely at mucosal surfaces. An immune correlates study demonstrated that antibodies targeting the V2 region and in a secondary analysis antibody-dependent cellular cytotoxicity (ADCC), in the presence of low envelope-specific (Env-specific) IgA, correlated with decreased risk of infection. Thus, understanding the B cell repertoires induced by this vaccine in systemic and mucosal compartments are key to understanding the potential protective mechanisms of this vaccine regimen. We immunized rhesus macaques with the ALVAC/AIDSVAX B/E gp120 vaccine regimen given in RV144, and then gave a boost 6 months later, after which the animals were necropsied. We isolated systemic and intestinal vaccine Env-specific memory B cells. Whereas Env-specific B cell clonal lineages were shared between spleen, draining inguinal, anterior pelvic, posterior pelvic, and periaortic lymph nodes, members of Env‑specific B cell clonal lineages were absent in the terminal ileum. Env‑specific antibodies were detectable in rectal fluids, suggesting that IgG antibodies present at mucosal sites were likely systemically produced and transported to intestinal mucosal sites.
Conflict of interest statement
J. Tartaglia was an employee of Sanofi and D. Francis, F. Sinangil, and C. Lee were employees of Global Solutions for Infectious Diseases at the time this study was performed. D. Francis, F. Sinangil, and C. Lee are former employees of VaxGen.
Figures



Similar articles
-
Characterization of HIV-1 gp120 antibody specificities induced in anogenital secretions of RV144 vaccine recipients after late boost immunizations.PLoS One. 2018 Apr 27;13(4):e0196397. doi: 10.1371/journal.pone.0196397. eCollection 2018. PLoS One. 2018. PMID: 29702672 Free PMC article. Clinical Trial.
-
Combined HIV-1 Envelope Systemic and Mucosal Immunization of Lactating Rhesus Monkeys Induces a Robust Immunoglobulin A Isotype B Cell Response in Breast Milk.J Virol. 2016 Apr 29;90(10):4951-4965. doi: 10.1128/JVI.00335-16. Print 2016 May 15. J Virol. 2016. PMID: 26937027 Free PMC article.
-
Boosting of HIV envelope CD4 binding site antibodies with long variable heavy third complementarity determining region in the randomized double blind RV305 HIV-1 vaccine trial.PLoS Pathog. 2017 Feb 24;13(2):e1006182. doi: 10.1371/journal.ppat.1006182. eCollection 2017 Feb. PLoS Pathog. 2017. PMID: 28235027 Free PMC article. Clinical Trial.
-
Lessons from the RV144 Thai phase III HIV-1 vaccine trial and the search for correlates of protection.Annu Rev Med. 2015;66:423-37. doi: 10.1146/annurev-med-052912-123749. Epub 2014 Oct 17. Annu Rev Med. 2015. PMID: 25341006 Review.
-
Prospects for a Globally Effective HIV-1 Vaccine.Am J Prev Med. 2015 Dec;49(6 Suppl 4):S307-18. doi: 10.1016/j.amepre.2015.09.004. Am J Prev Med. 2015. PMID: 26590431 Review.
Cited by
-
Bridging Vaccine-Induced HIV-1 Neutralizing and Effector Antibody Responses in Rabbit and Rhesus Macaque Animal Models.J Virol. 2019 May 1;93(10):e02119-18. doi: 10.1128/JVI.02119-18. Print 2019 May 15. J Virol. 2019. PMID: 30842326 Free PMC article.
-
Extracellular Loops of the Treponema pallidum FadL Orthologs TP0856 and TP0858 Elicit IgG Antibodies and IgG+-Specific B-Cells in the Rabbit Model of Experimental Syphilis.mBio. 2022 Aug 30;13(4):e0163922. doi: 10.1128/mbio.01639-22. Epub 2022 Jul 12. mBio. 2022. PMID: 35862766 Free PMC article.
-
Multivariate analysis of FcR-mediated NK cell functions identifies unique clustering among humans and rhesus macaques.Front Immunol. 2023 Dec 6;14:1260377. doi: 10.3389/fimmu.2023.1260377. eCollection 2023. Front Immunol. 2023. PMID: 38124734 Free PMC article.
-
Different adjuvanted pediatric HIV envelope vaccines induced distinct plasma antibody responses despite similar B cell receptor repertoires in infant rhesus macaques.PLoS One. 2021 Dec 31;16(12):e0256885. doi: 10.1371/journal.pone.0256885. eCollection 2021. PLoS One. 2021. PMID: 34972105 Free PMC article.
-
Application of area scaling analysis to identify natural killer cell and monocyte involvement in the GranToxiLux antibody dependent cell-mediated cytotoxicity assay.Cytometry A. 2018 Apr;93(4):436-447. doi: 10.1002/cyto.a.23348. Epub 2018 Mar 2. Cytometry A. 2018. PMID: 29498807 Free PMC article. Clinical Trial.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

