The Dynamic Interplay Between DNA Topoisomerases and DNA Topology
- PMID: 27942270
- PMCID: PMC5145260
- DOI: 10.1007/s12551-016-0206-x
The Dynamic Interplay Between DNA Topoisomerases and DNA Topology
Abstract
Topological properties of DNA influence its structure and biochemical interactions. Within the cell DNA topology is constantly in flux. Transcription and other essential processes including DNA replication and repair, alter the topology of the genome, while introducing additional complications associated with DNA knotting and catenation. These topological perturbations are counteracted by the action of topoisomerases, a specialized class of highly conserved and essential enzymes that actively regulate the topological state of the genome. This dynamic interplay among DNA topology, DNA processing enzymes, and DNA topoisomerases, is a pervasive factor that influences DNA metabolism in vivo. Building on the extensive structural and biochemical characterization over the past four decades that established the fundamental mechanistic basis of topoisomerase activity, the unique roles played by DNA topology in modulating and influencing the activity of topoisomerases have begun to be explored. In this review we survey established and emerging DNA topology dependent protein-DNA interactions with a focus on in vitro measurements of the dynamic interplay between DNA topology and topoisomerase activity.
Keywords: DNA supercoiling; DNA topology; cancer therapy; protein; topoisomerase.
Conflict of interest statement
Conflict of interest
Yeonee Seol declares that she has no conflict of interest.
Keir C. Neuman declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Figures
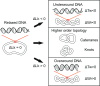

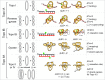
Similar articles
-
The dynamic interplay between DNA topoisomerases and DNA topology.Biophys Rev. 2016 Nov;8(Suppl 1):101-111. doi: 10.1007/s12551-016-0240-8. Epub 2016 Nov 14. Biophys Rev. 2016. PMID: 28510219 Free PMC article. Review.
-
The Coordinated Positive Regulation of Topoisomerase Genes Maintains Topological Homeostasis in Streptomyces coelicolor.J Bacteriol. 2016 Oct 7;198(21):3016-3028. doi: 10.1128/JB.00530-16. Print 2016 Nov 1. J Bacteriol. 2016. PMID: 27551021 Free PMC article.
-
DNA-Topology Simplification by Topoisomerases.Molecules. 2021 Jun 3;26(11):3375. doi: 10.3390/molecules26113375. Molecules. 2021. PMID: 34204901 Free PMC article. Review.
-
The benefit of DNA supercoiling during replication.Biochem Soc Trans. 2013 Apr;41(2):646-51. doi: 10.1042/BST20120281. Biochem Soc Trans. 2013. PMID: 23514170 Review.
-
DNA topoisomerases: structure, function, and mechanism.Annu Rev Biochem. 2001;70:369-413. doi: 10.1146/annurev.biochem.70.1.369. Annu Rev Biochem. 2001. PMID: 11395412 Review.
Cited by
-
A highly processive actinobacterial topoisomerase I - thoughts on Streptomyces' demand for an enzyme with a unique C-terminal domain.Microbiology (Reading). 2020 Feb;166(2):120-128. doi: 10.1099/mic.0.000841. Epub 2019 Aug 7. Microbiology (Reading). 2020. PMID: 31390324 Free PMC article. Review.
-
Supercoiling-dependent DNA binding: quantitative modeling and applications to bulk and single-molecule experiments.Nucleic Acids Res. 2024 Jan 11;52(1):59-72. doi: 10.1093/nar/gkad1055. Nucleic Acids Res. 2024. PMID: 38000393 Free PMC article.
-
Allele-specific collateral and fitness effects determine the dynamics of fluoroquinolone resistance evolution.Proc Natl Acad Sci U S A. 2022 May 3;119(18):e2121768119. doi: 10.1073/pnas.2121768119. Epub 2022 Apr 27. Proc Natl Acad Sci U S A. 2022. PMID: 35476512 Free PMC article.
-
The hyperthermophilic archaeon Thermococcus kodakarensis is resistant to pervasive negative supercoiling activity of DNA gyrase.Nucleic Acids Res. 2021 Dec 2;49(21):12332-12347. doi: 10.1093/nar/gkab869. Nucleic Acids Res. 2021. PMID: 34755863 Free PMC article.
-
Topoisomerases and cancer chemotherapy: recent advances and unanswered questions.F1000Res. 2019 Sep 30;8:F1000 Faculty Rev-1704. doi: 10.12688/f1000research.20201.1. eCollection 2019. F1000Res. 2019. PMID: 31602296 Free PMC article. Review.
References
-
- Bancaud A, Conde e Silva N, Barbi M, et al (2006) Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat Struct Mol Biol 13:444–450. http://www.nature.com/nsmb/journal/v13/n5/suppinfo/nsmb1087_S1.html - PubMed
-
- Bates AD, Maxwell A. DNA topology. 2. New York: Oxford University Press; 2005.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

