Relationships and Evolution of Double-Stranded RNA Totiviruses of Yeasts Inferred from Analysis of L-A-2 and L-BC Variants in Wine Yeast Strain Populations
- PMID: 27940540
- PMCID: PMC5288835
- DOI: 10.1128/AEM.02991-16
Relationships and Evolution of Double-Stranded RNA Totiviruses of Yeasts Inferred from Analysis of L-A-2 and L-BC Variants in Wine Yeast Strain Populations
Abstract
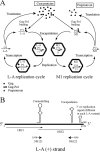
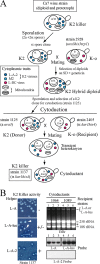
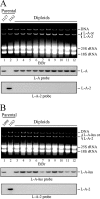
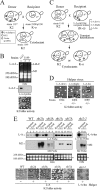
Saccharomyces cerevisiae killer strains secrete a protein toxin active on nonkiller strains of the same (or other) yeast species. Different killer toxins, K1, K2, K28, and Klus, have been described. Each toxin is encoded by a medium-size (1.5- to 2.3-kb) M double-stranded RNA (dsRNA) located in the cytoplasm. M dsRNAs require L-A helper virus for maintenance. L-A belongs to the Totiviridae family, and its dsRNA genome of 4.6 kb codes for the major capsid protein Gag and a minor Gag-Pol protein, which form the virions that separately encapsidate L-A or the M satellites. Different L-A variants exist in nature; on average, 24% of their nucleotides are different. Previously, we reported that L-A-lus was specifically associated with Mlus, suggesting coevolution, and proposed a role of the toxin-encoding M dsRNAs in the appearance of new L-A variants. Here we confirm this by analyzing the helper virus in K2 killer wine strains, which we named L-A-2. L-A-2 is required for M2 maintenance, and neither L-A nor L-A-lus shows helper activity for M2 in the same genetic background. This requirement is overcome when coat proteins are provided in large amounts by a vector or in ski mutants. The genome of another totivirus, L-BC, frequently accompanying L-A in the same cells shows a lower degree of variation than does L-A (about 10% of nucleotides are different). Although L-BC has no helper activity for M dsRNAs, distinct L-BC variants are associated with a particular killer strain. The so-called L-BC-lus (in Klus strains) and L-BC-2 (in K2 strains) are analyzed.
Importance: Killer strains of S. cerevisiae secrete protein toxins that kill nonkiller yeasts. The "killer phenomenon" depends on two dsRNA viruses: L-A and M. M encodes the toxin, and L-A, the helper virus, provides the capsids for both viruses. Different killer toxins exist: K1, K2, K28, and Klus, encoded on different M viruses. Our data indicate that each M dsRNA depends on a specific helper virus; these helper viruses have nucleotide sequences that may be as much as 26% different, suggesting coevolution. In wine environments, K2 and Klus strains frequently coexist. We have previously characterized the association of Mlus and L-A-lus. Here we sequence and characterize L-A-2, the helper virus of M2, establishing the helper virus requirements of M2, which had not been completely elucidated. We also report the existence of two specific L-BC totiviruses in Klus and K2 strains with about 10% of their nucleotides different, suggesting different evolutionary histories from those of L-A viruses.
Keywords: L-A helper virus; double-stranded RNA virus; yeast killer toxins; yeast virus; yeast wine strains.
Copyright © 2017 American Society for Microbiology.
Figures

Similar articles
-
L-A-lus, a new variant of the L-A totivirus found in wine yeasts with Klus killer toxin-encoding Mlus double-stranded RNA: possible role of killer toxin-encoding satellite RNAs in the evolution of their helper viruses.Appl Environ Microbiol. 2013 Aug;79(15):4661-74. doi: 10.1128/AEM.00500-13. Epub 2013 May 31. Appl Environ Microbiol. 2013. PMID: 23728812 Free PMC article.
-
A Simple Multiplex Reverse Transcription-PCR Method for the Diagnosis of L-A and M Totiviruses in Saccharomyces cerevisiae.Appl Environ Microbiol. 2022 Feb 22;88(4):e0221321. doi: 10.1128/AEM.02213-21. Epub 2021 Dec 15. Appl Environ Microbiol. 2022. PMID: 34910561 Free PMC article.
-
Variation and Distribution of L-A Helper Totiviruses in Saccharomyces sensu stricto Yeasts Producing Different Killer Toxins.Toxins (Basel). 2017 Oct 11;9(10):313. doi: 10.3390/toxins9100313. Toxins (Basel). 2017. PMID: 29019944 Free PMC article.
-
Double-stranded and single-stranded RNA viruses of Saccharomyces cerevisiae.Annu Rev Microbiol. 1992;46:347-75. doi: 10.1146/annurev.mi.46.100192.002023. Annu Rev Microbiol. 1992. PMID: 1444259 Review.
-
Yeast Killer Toxin K28: Biology and Unique Strategy of Host Cell Intoxication and Killing.Toxins (Basel). 2017 Oct 20;9(10):333. doi: 10.3390/toxins9100333. Toxins (Basel). 2017. PMID: 29053588 Free PMC article. Review.
Cited by
-
Specificity Determination in Saccharomyces cerevisiae Killer Virus Systems.Microorganisms. 2021 Jan 23;9(2):236. doi: 10.3390/microorganisms9020236. Microorganisms. 2021. PMID: 33498746 Free PMC article.
-
Expression of the K74 Killer Toxin from Saccharomyces paradoxus Is Modulated by the Toxin-Encoding M74 Double-Stranded RNA 5' Untranslated Terminal Region.Appl Environ Microbiol. 2022 Apr 26;88(8):e0203021. doi: 10.1128/aem.02030-21. Epub 2022 Apr 7. Appl Environ Microbiol. 2022. PMID: 35389250 Free PMC article.
-
Genome Organization of a New Double-Stranded RNA LA Helper Virus From Wine Torulaspora delbrueckii Killer Yeast as Compared With Its Saccharomyces Counterparts.Front Microbiol. 2020 Nov 23;11:593846. doi: 10.3389/fmicb.2020.593846. eCollection 2020. Front Microbiol. 2020. PMID: 33324373 Free PMC article.
-
Dynamic modelling of the killing mechanism of action by virus-infected yeasts.J R Soc Interface. 2019 Mar 29;16(152):20190064. doi: 10.1098/rsif.2019.0064. J R Soc Interface. 2019. PMID: 30890050 Free PMC article.
-
Evaluating Atlantic Salmon (Salmo salar) as a Natural or Alternative Host for Piscine Myocarditis Virus (PMCV) Infection.Pathogens. 2024 Aug 30;13(9):744. doi: 10.3390/pathogens13090744. Pathogens. 2024. PMID: 39338935 Free PMC article.
References
-
- Rodríguez-Cousiño N, Maqueda M, Ambrona J, Zamora E, Esteban R, Ramírez M. 2011. A new wine Saccharomyces cerevisiae killer toxin (Klus), encoded by a double-stranded RNA virus, with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl Environ Microbiol 77:1822–1832. doi:10.1128/AEM.02501-10. - DOI - PMC - PubMed
-
- Icho T, Wickner RB. 1989. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J Biol Chem 264:6716–6723. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

