AvrPm2 encodes an RNase-like avirulence effector which is conserved in the two different specialized forms of wheat and rye powdery mildew fungus
- PMID: 27935041
- PMCID: PMC5347869
- DOI: 10.1111/nph.14372
AvrPm2 encodes an RNase-like avirulence effector which is conserved in the two different specialized forms of wheat and rye powdery mildew fungus
Erratum in
-
Corrigendum.New Phytol. 2019 Jun;222(4):2038. doi: 10.1111/nph.15831. Epub 2019 Apr 10. New Phytol. 2019. PMID: 31066073 Free PMC article. No abstract available.
Abstract
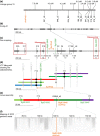
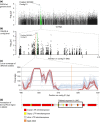
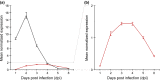
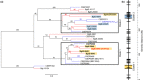
There is a large diversity of genetically defined resistance genes in bread wheat against the powdery mildew pathogen Blumeria graminis (B. g.) f. sp. tritici. Many confer race-specific resistance to this pathogen, but until now only the mildew avirulence gene AvrPm3a2/f2 that is recognized by Pm3a/f was known molecularly. We performed map-based cloning and genome-wide association studies to isolate a candidate for the mildew avirulence gene AvrPm2. We then used transient expression assays in Nicotiana benthamiana to demonstrate specific and strong recognition of AvrPm2 by Pm2. The virulent AvrPm2 allele arose from a conserved 12 kb deletion, while there is no protein sequence diversity in the gene pool of avirulent B. g. tritici isolates. We found one polymorphic AvrPm2 allele in B. g. triticale and one orthologue in B. g. secalis and both are recognized by Pm2. AvrPm2 belongs to a small gene family encoding structurally conserved RNase-like effectors, including Avra13 from B. g. hordei, the cognate Avr of the barley resistance gene Mla13. These results demonstrate the conservation of functional avirulence genes in two cereal powdery mildews specialized on different hosts, thus providing a possible explanation for successful introgression of resistance genes from rye or other grass relatives to wheat.
Keywords: Blumeria graminis; Pm2; RNAse-like; avirulence gene; powdery mildew; wheat.
© 2016 The Authors. New Phytologist © 2016 New Phytologist Trust.
Figures


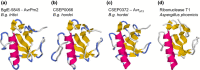
Comment in
-
Cereal immunity against powdery mildews targets RNase-Like Proteins associated with Haustoria (RALPH) effectors evolved from a common ancestral gene.New Phytol. 2017 Feb;213(3):969-971. doi: 10.1111/nph.14386. New Phytol. 2017. PMID: 28079937 No abstract available.
Similar articles
-
Identification of specificity-defining amino acids of the wheat immune receptor Pm2 and powdery mildew effector AvrPm2.Plant J. 2021 May;106(4):993-1007. doi: 10.1111/tpj.15214. Epub 2021 Apr 8. Plant J. 2021. PMID: 33629439
-
Distinct domains of the AVRPM3A2/F2 avirulence protein from wheat powdery mildew are involved in immune receptor recognition and putative effector function.New Phytol. 2018 Apr;218(2):681-695. doi: 10.1111/nph.15026. Epub 2018 Feb 17. New Phytol. 2018. PMID: 29453934 Free PMC article.
-
Multiple Avirulence Loci and Allele-Specific Effector Recognition Control the Pm3 Race-Specific Resistance of Wheat to Powdery Mildew.Plant Cell. 2015 Oct;27(10):2991-3012. doi: 10.1105/tpc.15.00171. Epub 2015 Oct 9. Plant Cell. 2015. PMID: 26452600 Free PMC article.
-
Cereal powdery mildew effectors: a complex toolbox for an obligate pathogen.Curr Opin Microbiol. 2018 Dec;46:26-33. doi: 10.1016/j.mib.2018.01.018. Epub 2018 Feb 22. Curr Opin Microbiol. 2018. PMID: 29455142 Review.
-
Long-term and rapid evolution in powdery mildew fungi.Mol Ecol. 2024 May;33(10):e16909. doi: 10.1111/mec.16909. Epub 2023 Mar 20. Mol Ecol. 2024. PMID: 36862075 Review.
Cited by
-
The Parauncinula polyspora Draft Genome Provides Insights into Patterns of Gene Erosion and Genome Expansion in Powdery Mildew Fungi.mBio. 2019 Sep 24;10(5):e01692-19. doi: 10.1128/mBio.01692-19. mBio. 2019. PMID: 31551331 Free PMC article.
-
Non-parent of Origin Expression of Numerous Effector Genes Indicates a Role of Gene Regulation in Host Adaption of the Hybrid Triticale Powdery Mildew Pathogen.Front Plant Sci. 2018 Jan 30;9:49. doi: 10.3389/fpls.2018.00049. eCollection 2018. Front Plant Sci. 2018. PMID: 29441081 Free PMC article.
-
Remote homology clustering identifies lowly conserved families of effector proteins in plant-pathogenic fungi.Microb Genom. 2021 Sep;7(9):000637. doi: 10.1099/mgen.0.000637. Microb Genom. 2021. PMID: 34468307 Free PMC article.
-
Comparative genome analyses reveal sequence features reflecting distinct modes of host-adaptation between dicot and monocot powdery mildew.BMC Genomics. 2018 Sep 25;19(1):705. doi: 10.1186/s12864-018-5069-z. BMC Genomics. 2018. PMID: 30253736 Free PMC article.
-
Structural polymorphisms within a common powdery mildew effector scaffold as a driver of coevolution with cereal immune receptors.Proc Natl Acad Sci U S A. 2023 Aug 8;120(32):e2307604120. doi: 10.1073/pnas.2307604120. Epub 2023 Jul 31. Proc Natl Acad Sci U S A. 2023. PMID: 37523523 Free PMC article.
References
-
- Bourras S, McNally KE, Ben‐David R, Parlange F, Roffler S, Praz CR, Oberhaensli S, Menardo F, Stirnweis D, Frenkel Z et al 2015. Multiple avirulence loci and allele‐specific effector recognition control the Pm3 race‐specific resistance of wheat to powdery mildew. Plant Cell 27: 2991–3012. - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

