Nup100 regulates Saccharomyces cerevisiae replicative life span by mediating the nuclear export of specific tRNAs
- PMID: 27932586
- PMCID: PMC5311497
- DOI: 10.1261/rna.057612.116
Nup100 regulates Saccharomyces cerevisiae replicative life span by mediating the nuclear export of specific tRNAs
Abstract
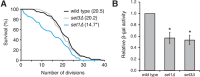
Nuclear pore complexes (NPCs), which are composed of nucleoporins (Nups) and regulate transport between the nucleus and cytoplasm, significantly impact the replicative life span (RLS) of Saccharomyces cerevisiae We previously reported that deletion of the nonessential gene NUP100 increases RLS, although the molecular basis for this effect was unknown. In this study, we find that nuclear tRNA accumulation contributes to increased longevity in nup100Δ cells. Fluorescence in situ hybridization (FISH) experiments demonstrate that several specific tRNAs accumulate in the nuclei of nup100Δ mutants. Protein levels of the transcription factor Gcn4 are increased when NUP100 is deleted, and GCN4 is required for the elevated life spans of nup100Δ mutants, similar to other previously described tRNA export and ribosomal mutants. Northern blots indicate that tRNA splicing and aminoacylation are not significantly affected in nup100Δ cells, suggesting that Nup100 is largely required for nuclear export of mature, processed tRNAs. Distinct tRNAs accumulate in the nuclei of nup100Δ and msn5Δ mutants, while Los1-GFP nucleocytoplasmic shuttling is unaffected by Nup100. Thus, we conclude that Nup100 regulates tRNA export in a manner distinct from Los1 or Msn5. Together, these experiments reveal a novel Nup100 role in the tRNA life cycle that impacts the S. cerevisiae life span.
Keywords: aging; nuclear pore complex; tRNA.
© 2017 Lord et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



Similar articles
-
A subset of FG-nucleoporins is necessary for efficient Msn5-mediated nuclear protein export.Biochim Biophys Acta. 2013 May;1833(5):1096-103. doi: 10.1016/j.bbamcr.2012.12.020. Epub 2013 Jan 4. Biochim Biophys Acta. 2013. PMID: 23295456 Free PMC article.
-
Regulation of tRNA bidirectional nuclear-cytoplasmic trafficking in Saccharomyces cerevisiae.Mol Biol Cell. 2010 Feb 15;21(4):639-49. doi: 10.1091/mbc.e09-07-0551. Epub 2009 Dec 23. Mol Biol Cell. 2010. PMID: 20032305 Free PMC article.
-
Utp9p facilitates Msn5p-mediated nuclear reexport of retrograded tRNAs in Saccharomyces cerevisiae.Mol Biol Cell. 2009 Dec;20(23):5007-25. doi: 10.1091/mbc.e09-06-0490. Epub 2009 Oct 7. Mol Biol Cell. 2009. PMID: 19812255 Free PMC article.
-
The ins and outs of nuclear re-export of retrogradely transported tRNAs in Saccharomyces cerevisiae.Nucleus. 2010 May-Jun;1(3):224-30. doi: 10.4161/nucl.1.3.11250. Epub 2010 Jan 13. Nucleus. 2010. PMID: 21327067 Free PMC article. Review.
-
Review: transport of tRNA out of the nucleus-direct channeling to the ribosome?J Struct Biol. 2000 Apr;129(2-3):288-94. doi: 10.1006/jsbi.2000.4226. J Struct Biol. 2000. PMID: 10806079 Review.
Cited by
-
Is Gcn4-induced autophagy the ultimate downstream mechanism by which hormesis extends yeast replicative lifespan?Curr Genet. 2019 Jun;65(3):717-720. doi: 10.1007/s00294-019-00936-4. Epub 2019 Jan 23. Curr Genet. 2019. PMID: 30673825 Free PMC article. Review.
-
Nuclear envelope-vacuole contacts mitigate nuclear pore complex assembly stress.J Cell Biol. 2020 Dec 7;219(12):e202001165. doi: 10.1083/jcb.202001165. J Cell Biol. 2020. PMID: 33053148 Free PMC article.
-
Poor old pores-The challenge of making and maintaining nuclear pore complexes in aging.FEBS J. 2020 Mar;287(6):1058-1075. doi: 10.1111/febs.15205. Epub 2020 Jan 23. FEBS J. 2020. PMID: 31912972 Free PMC article. Review.
-
Karyopherin Msn5 is involved in a novel mechanism controlling the cellular level of cell cycle regulators Cln2 and Swi5.Cell Cycle. 2019 Mar;18(5):580-595. doi: 10.1080/15384101.2019.1578148. Epub 2019 Feb 11. Cell Cycle. 2019. PMID: 30739521 Free PMC article.
-
A nuclear pore sub-complex restricts the propagation of Ty retrotransposons by limiting their transcription.PLoS Genet. 2021 Nov 1;17(11):e1009889. doi: 10.1371/journal.pgen.1009889. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34723966 Free PMC article.
References
-
- Dever TE. 1997. Using GCN4 as a reporter of eIF2α phosphorylation and translational regulation in yeast. Methods 11: 403–417. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
