A Near-Atomic Structure of the Dark Apoptosome Provides Insight into Assembly and Activation
- PMID: 27916517
- PMCID: PMC5214966
- DOI: 10.1016/j.str.2016.11.002
A Near-Atomic Structure of the Dark Apoptosome Provides Insight into Assembly and Activation
Abstract
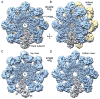
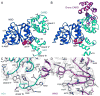
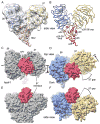
In Drosophila, the Apaf-1-related killer (Dark) forms an apoptosome that activates procaspases. To investigate function, we have determined a near-atomic structure of Dark double rings using cryo-electron microscopy. We then built a nearly complete model of the apoptosome that includes 7- and 8-blade β-propellers. We find that the preference for dATP during Dark assembly may be governed by Ser325, which is in close proximity to the 2' carbon of the deoxyribose ring. Interestingly, β-propellers in V-shaped domains of the Dark apoptosome are more widely separated, relative to these features in the Apaf-1 apoptosome. This wider spacing may be responsible for the lack of cytochrome c binding to β-propellers in the Dark apoptosome. Our structure also highlights the roles of two loss-of-function mutations that may block Dark assembly. Finally, the improved model provides a framework to understand apical procaspase activation in the intrinsic cell death pathway.
Keywords: apoptosis; apoptosome; procaspase activation; programmed cell death; single particle cryo-EM.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures





Similar articles
-
New insights into apoptosome structure and function.Cell Death Differ. 2018 Jul;25(7):1194-1208. doi: 10.1038/s41418-017-0025-z. Epub 2018 May 15. Cell Death Differ. 2018. PMID: 29765111 Free PMC article. Review.
-
Structure of the Drosophila apoptosome at 6.9 å resolution.Structure. 2011 Jan 12;19(1):128-40. doi: 10.1016/j.str.2010.10.009. Structure. 2011. PMID: 21220123 Free PMC article.
-
Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila.Genes Dev. 2015 Feb 1;29(3):277-87. doi: 10.1101/gad.255877.114. Genes Dev. 2015. PMID: 25644603 Free PMC article.
-
Apoptosome assembly.Methods Enzymol. 2008;442:141-56. doi: 10.1016/S0076-6879(08)01407-9. Methods Enzymol. 2008. PMID: 18662568
-
Apoptosome structure, assembly, and procaspase activation.Structure. 2013 Apr 2;21(4):501-15. doi: 10.1016/j.str.2013.02.024. Structure. 2013. PMID: 23561633 Free PMC article. Review.
Cited by
-
NLR immune receptors: structure and function in plant disease resistance.Biochem Soc Trans. 2023 Aug 31;51(4):1473-1483. doi: 10.1042/BST20221087. Biochem Soc Trans. 2023. PMID: 37602488 Free PMC article. Review.
-
New insights into apoptosome structure and function.Cell Death Differ. 2018 Jul;25(7):1194-1208. doi: 10.1038/s41418-017-0025-z. Epub 2018 May 15. Cell Death Differ. 2018. PMID: 29765111 Free PMC article. Review.
-
Non-apoptotic enteroblast-specific role of the initiator caspase Dronc for development and homeostasis of the Drosophila intestine.Sci Rep. 2021 Jan 29;11(1):2645. doi: 10.1038/s41598-021-81261-0. Sci Rep. 2021. PMID: 33514791 Free PMC article.
-
Proapoptotic function of deubiquitinase DUSP31 in Drosophila.Oncotarget. 2017 Jul 31;8(41):70452-70462. doi: 10.18632/oncotarget.19715. eCollection 2017 Sep 19. Oncotarget. 2017. PMID: 29050293 Free PMC article.
-
Diversity in the intrinsic apoptosis pathway of nematodes.Commun Biol. 2020 Aug 28;3(1):478. doi: 10.1038/s42003-020-01208-5. Commun Biol. 2020. PMID: 32859965 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. - PMC - PubMed
-
- Akdemir F, Farkas R, Chen P, Juhasz G, Medved’ová L, Sass M, Wang L, Wang X, Chittaranjan S, Gorski SM, Rodriguez A, Abrams JM. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006;133:1457–1465. - PubMed
-
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS. A unified model for apical caspase activation. Molecular Cell. 2003;11:529–541. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

