The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease
- PMID: 27911701
- PMCID: PMC5095923
- DOI: 10.1042/BST20160172
The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease
Abstract
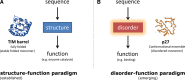
In the 1960s, Christian Anfinsen postulated that the unique three-dimensional structure of a protein is determined by its amino acid sequence. This work laid the foundation for the sequence-structure-function paradigm, which states that the sequence of a protein determines its structure, and structure determines function. However, a class of polypeptide segments called intrinsically disordered regions does not conform to this postulate. In this review, I will first describe established and emerging ideas about how disordered regions contribute to protein function. I will then discuss molecular principles by which regulatory mechanisms, such as alternative splicing and asymmetric localization of transcripts that encode disordered regions, can increase the functional versatility of proteins. Finally, I will discuss how disordered regions contribute to human disease and the emergence of cellular complexity during organismal evolution.
Keywords: RNA localization; alternative splicing; biological networks; gene expression and regulation; intrinsically disordered proteins; protein turnover.
© 2016 The Author(s).
Figures


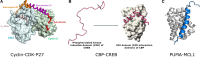





Similar articles
-
Alternative splicing of intrinsically disordered regions and rewiring of protein interactions.Curr Opin Struct Biol. 2013 Jun;23(3):443-50. doi: 10.1016/j.sbi.2013.03.006. Epub 2013 May 22. Curr Opin Struct Biol. 2013. PMID: 23706950 Review.
-
Functions of short lifetime biological structures at large: the case of intrinsically disordered proteins.Brief Funct Genomics. 2020 Jan 22;19(1):60-68. doi: 10.1093/bfgp/ely023. Brief Funct Genomics. 2020. PMID: 29982297 Review.
-
The contribution of intrinsic disorder prediction to the elucidation of protein function.Curr Opin Struct Biol. 2013 Jun;23(3):467-72. doi: 10.1016/j.sbi.2013.02.001. Epub 2013 Mar 1. Curr Opin Struct Biol. 2013. PMID: 23466039 Review.
-
Expanding the Paradigm: Intrinsically Disordered Proteins and Allosteric Regulation.J Mol Biol. 2018 Aug 3;430(16):2309-2320. doi: 10.1016/j.jmb.2018.04.003. Epub 2018 Apr 7. J Mol Biol. 2018. PMID: 29634920 Free PMC article. Review.
-
Coupled binding and folding of intrinsically disordered proteins: what can we learn from kinetics?Curr Opin Struct Biol. 2016 Feb;36:18-24. doi: 10.1016/j.sbi.2015.11.012. Epub 2015 Dec 22. Curr Opin Struct Biol. 2016. PMID: 26720267 Review.
Cited by
-
NAP1L1 and NAP1L4 Binding to Hypervariable Domain of Chikungunya Virus nsP3 Protein Is Bivalent and Requires Phosphorylation.J Virol. 2021 Jul 26;95(16):e0083621. doi: 10.1128/JVI.00836-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34076483 Free PMC article.
-
The Long Linker Region of Telomere-Binding Protein TRF2 Is Responsible for Interactions with Lamins.Int J Mol Sci. 2021 Mar 24;22(7):3293. doi: 10.3390/ijms22073293. Int J Mol Sci. 2021. PMID: 33804854 Free PMC article.
-
Conservation of land plant-specific receptor-like cytoplasmic kinase subfamily XI possessing a unique kinase insert domain.Front Plant Sci. 2023 Feb 22;14:1117059. doi: 10.3389/fpls.2023.1117059. eCollection 2023. Front Plant Sci. 2023. PMID: 36909417 Free PMC article.
-
Features of molecular recognition of intrinsically disordered proteins via coupled folding and binding.Protein Sci. 2019 Nov;28(11):1952-1965. doi: 10.1002/pro.3718. Epub 2019 Sep 4. Protein Sci. 2019. PMID: 31441158 Free PMC article. Review.
-
Protein proximity networks and functional evaluation of the casein kinase 1 gamma family reveal unique roles for CK1γ3 in WNT signaling.J Biol Chem. 2022 Jun;298(6):101986. doi: 10.1016/j.jbc.2022.101986. Epub 2022 Apr 27. J Biol Chem. 2022. PMID: 35487243 Free PMC article.
References
-
- Romero P., Obradovic Z., Kissinger C.R., Villafranca J.E., Garner E., Guilliot S. et al. (1998) Thousands of proteins likely to have long disordered regions. Pac. Symp. Biocomput. 437–448 PMID: - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

