Infection of rhesus macaques with a pool of simian immunodeficiency virus with the envelope genes from acute HIV-1 infections
- PMID: 27906032
- PMCID: PMC5124249
- DOI: 10.1186/s12981-016-0125-8
Infection of rhesus macaques with a pool of simian immunodeficiency virus with the envelope genes from acute HIV-1 infections
Abstract
Background: New simian-human immunodeficiency chimeric viruses with an HIV-1 env (SHIVenv) are critical for studies on HIV pathogenesis, vaccine development, and microbicide testing. Macaques are typically exposed to single CCR5-using SHIVenv which in most instances does not reflect the conditions during acute/early HIV infection (AHI) in humans. Instead of individual and serial testing new SHIV constructs, a pool of SHIVenv_B derived from 16 acute HIV-1 infections were constructed using a novel yeast-based SHIV cloning approach and then used to infect macaques.
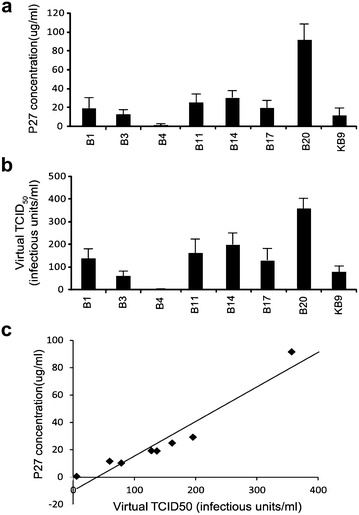
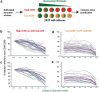
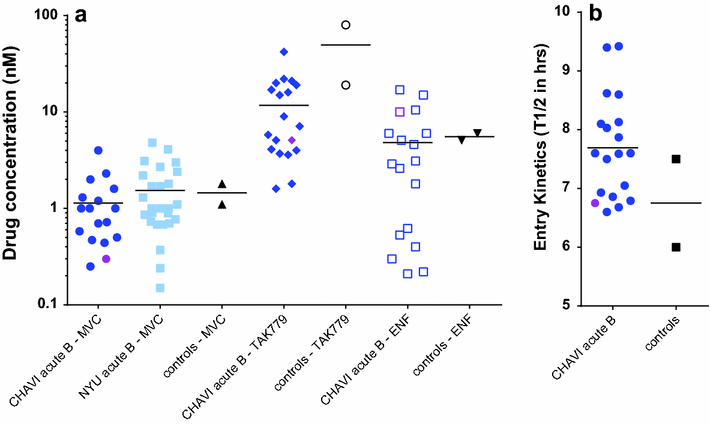
Results: Even though none of the 16 SHIVenvs contained the recently reported mutations in env genes that could significantly enhance their binding affinity to RhCD4, one SHIVenv (i.e. SHIVenv_B3-PRB926) established infection in macaques exposed to this pool. AHI SHIVenv_B viruses as well as their HIVenv_B counterparts were analyzed for viral protein content, function, and fitness to identify possible difference between SHIVenv_B3-PRB926 and the other 15 SHIVenvs in the pool. All of the constructs produced SHIV or HIV chimeric with wild type levels of capsid (p27 and p24) content, reverse transcriptase (RT) activity, and expressed envelope glycoproteins that could bind to cell receptors CD4/CCR5 and mediate virus entry. HIV-1env_B chimeric viruses were propagated in susceptible cell lines but the 16 SHIVenv_B variants showed only limited replication in macaque peripheral blood mononuclear cells (PBMCs) and 174×CEM.CCR5 cell line. AHI chimeric viruses including HIVenv_B3 showed only minor variations in cell entry efficiency and kinetics as well as replicative fitness in human PBMCs. Reduced number of N-link glycosylation sites and slightly greater CCR5 affinity/avidity was the only distinguishing feature of env_B3 versus other AHI env's in the pool, a feature also observed in the HIV establishing new infections in humans.
Conclusion: Despite the inability to propagate in primary cells and cell lines, a pool of 16 SHIVenv viruses could establish infection but only one virus, SHIVenv_B3 was isolated in the macaque and then shown to repeatedly infected macaques. This SHIVenv_B3 virus did not show any distinct phenotypic property from the other 15 SHIVenv viruses but did have the fewest N-linked glycosylation sites.
Keywords: Glycosylation; Replicative fitness; Simian–human immunodeficiency chimeric virus; Transmission.
Figures


Similar articles
-
Pathogenic infection of Rhesus macaques by an evolving SIV-HIV derived from CCR5-using envelope genes of acute HIV-1 infections.Virology. 2016 Dec;499:298-312. doi: 10.1016/j.virol.2016.09.021. Epub 2016 Oct 7. Virology. 2016. PMID: 27723488 Free PMC article.
-
Conformation of HIV-1 Envelope Governs Rhesus CD4 Usage and Simian-Human Immunodeficiency Virus Replication.mBio. 2022 Feb 22;13(1):e0275221. doi: 10.1128/mbio.02752-21. Epub 2022 Jan 11. mBio. 2022. PMID: 35012342 Free PMC article.
-
Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques.Proc Natl Acad Sci U S A. 2016 Jun 14;113(24):E3413-22. doi: 10.1073/pnas.1606636113. Epub 2016 May 31. Proc Natl Acad Sci U S A. 2016. PMID: 27247400 Free PMC article.
-
Understanding the basis of CD4(+) T-cell depletion in macaques infected by a simian-human immunodeficiency virus.Vaccine. 2002 May 6;20(15):1934-7. doi: 10.1016/s0264-410x(02)00072-5. Vaccine. 2002. PMID: 11983249 Review.
-
CD4-HIV-1 Envelope Interactions: Critical Insights for the Simian/HIV/Macaque Model.AIDS Res Hum Retroviruses. 2018 Sep;34(9):778-779. doi: 10.1089/AID.2018.0110. Epub 2018 Jul 9. AIDS Res Hum Retroviruses. 2018. PMID: 29886767 Free PMC article. Review.
Cited by
-
Deep Gene Sequence Cluster Analyses of Multi-Virus-Infected Mucosal Tissue Reveal Enhanced Transmission of Acute HIV-1.J Virol. 2021 Jan 13;95(3):e01737-20. doi: 10.1128/JVI.01737-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33177204 Free PMC article.
-
Pathogenic infection of Rhesus macaques by an evolving SIV-HIV derived from CCR5-using envelope genes of acute HIV-1 infections.Virology. 2016 Dec;499:298-312. doi: 10.1016/j.virol.2016.09.021. Epub 2016 Oct 7. Virology. 2016. PMID: 27723488 Free PMC article.
References
-
- Nishimura Y, Igarashi T, Donau OK, Buckler-White A, Buckler C, Lafont BA, et al. Highly pathogenic SHIVs and SIVs target different CD4+ T cell subsets in rhesus monkeys, explaining their divergent clinical courses. Proc Natl Acad Sci USA. 2004;101:12324–12329. doi: 10.1073/pnas.0404620101. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

