MeFiT: merging and filtering tool for illumina paired-end reads for 16S rRNA amplicon sequencing
- PMID: 27905885
- PMCID: PMC5134250
- DOI: 10.1186/s12859-016-1358-1
MeFiT: merging and filtering tool for illumina paired-end reads for 16S rRNA amplicon sequencing
Abstract
Background: Recent advances in next-generation sequencing have revolutionized genomic research. 16S rRNA amplicon sequencing using paired-end sequencing on the MiSeq platform from Illumina, Inc., is being used to characterize the composition and dynamics of extremely complex/diverse microbial communities. For this analysis on the Illumina platform, merging and quality filtering of paired-end reads are essential first steps in data analysis to ensure the accuracy and reliability of downstream analysis.
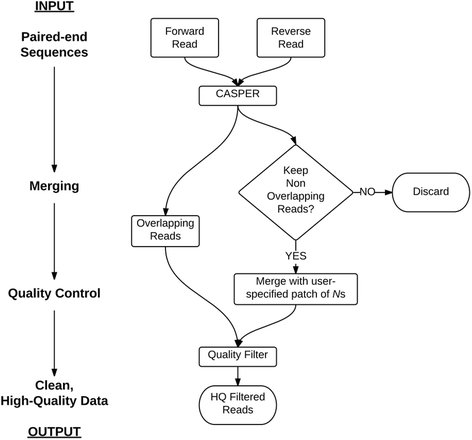
Results: We have developed the Merging and Filtering Tool (MeFiT) to combine these pre-processing steps into one simple, intuitive pipeline. MeFiT invokes CASPER (context-aware scheme for paired-end reads) for merging paired-end reads and provides users the option to quality filter the reads using the traditional average Q-score metric or using a maximum expected error cut-off threshold.
Conclusions: MeFiT provides an open-source solution that permits users to merge and filter paired end illumina reads. The tool has been implemented in python and the source-code is freely available at https://github.com/nisheth/MeFiT .
Keywords: 16S rRNA; microbiome; paired-end sequencing; pre-processing; quality filtering.
Similar articles
-
IPED: a highly efficient denoising tool for Illumina MiSeq Paired-end 16S rRNA gene amplicon sequencing data.BMC Bioinformatics. 2016 Apr 29;17(1):192. doi: 10.1186/s12859-016-1061-2. BMC Bioinformatics. 2016. PMID: 27130479 Free PMC article.
-
CASPER: context-aware scheme for paired-end reads from high-throughput amplicon sequencing.BMC Bioinformatics. 2014;15 Suppl 9(Suppl 9):S10. doi: 10.1186/1471-2105-15-S9-S10. Epub 2014 Sep 10. BMC Bioinformatics. 2014. PMID: 25252785 Free PMC article.
-
Evaluation of 16S rRNA amplicon sequencing using two next-generation sequencing technologies for phylogenetic analysis of the rumen bacterial community in steers.J Microbiol Methods. 2016 Aug;127:132-140. doi: 10.1016/j.mimet.2016.06.004. Epub 2016 Jun 6. J Microbiol Methods. 2016. PMID: 27282101
-
Generating amplicon reads for microbial community assessment with next-generation sequencing.J Appl Microbiol. 2020 Feb;128(2):330-354. doi: 10.1111/jam.14380. Epub 2019 Aug 1. J Appl Microbiol. 2020. PMID: 31299126 Review.
-
MiSeq: A Next Generation Sequencing Platform for Genomic Analysis.Methods Mol Biol. 2018;1706:223-232. doi: 10.1007/978-1-4939-7471-9_12. Methods Mol Biol. 2018. PMID: 29423801 Review.
Cited by
-
Microbiome data reveal significant differences in the bacterial diversity in freshwater rohu (Labeo rohita) across the supply chain in Dhaka, Bangladesh.Lett Appl Microbiol. 2022 Oct;75(4):813-823. doi: 10.1111/lam.13739. Epub 2022 Jun 13. Lett Appl Microbiol. 2022. PMID: 35575585 Free PMC article.
-
Race, the Vaginal Microbiome, and Spontaneous Preterm Birth.mSystems. 2022 Jun 28;7(3):e0001722. doi: 10.1128/msystems.00017-22. Epub 2022 May 18. mSystems. 2022. PMID: 35582911 Free PMC article.
-
Efficacy and safety of a food supplement with standardized menthol, limonene, and gingerol content in patients with irritable bowel syndrome: A double-blind, randomized, placebo-controlled trial.PLoS One. 2022 Jun 15;17(6):e0263880. doi: 10.1371/journal.pone.0263880. eCollection 2022. PLoS One. 2022. PMID: 35704960 Free PMC article. Clinical Trial.
-
Compositional Bias in Naïve and Chemically-modified Phage-Displayed Libraries uncovered by Paired-end Deep Sequencing.Sci Rep. 2018 Jan 19;8(1):1214. doi: 10.1038/s41598-018-19439-2. Sci Rep. 2018. PMID: 29352178 Free PMC article.
-
ITS and 16S rDNA metagenomic dataset of different soils from flax fields.Data Brief. 2023 Nov 18;52:109827. doi: 10.1016/j.dib.2023.109827. eCollection 2024 Feb. Data Brief. 2023. PMID: 38059001 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources