The Human Cytomegalovirus IE1 Protein Antagonizes PML Nuclear Body-Mediated Intrinsic Immunity via the Inhibition of PML De Novo SUMOylation
- PMID: 27903803
- PMCID: PMC5286881
- DOI: 10.1128/JVI.02049-16
The Human Cytomegalovirus IE1 Protein Antagonizes PML Nuclear Body-Mediated Intrinsic Immunity via the Inhibition of PML De Novo SUMOylation
Abstract
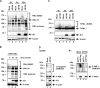
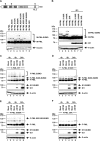
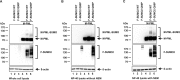
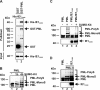
PML nuclear bodies (NBs) are accumulations of cellular proteins embedded in a scaffold-like structure built by SUMO-modified PML/TRIM19. PML and other NB proteins act as cellular restriction factors against human cytomegalovirus (HCMV); however, this intrinsic defense is counteracted by the immediate early protein 1 (IE1) of HCMV. IE1 directly interacts with the PML coiled-coil domain via its globular core region and disrupts NB foci by inducing a loss of PML SUMOylation. Here, we demonstrate that IE1 acts via abrogating the de novo SUMOylation of PML. In order to overcome reversible SUMOylation dynamics, we made use of a cell-based assay that combines inducible IE1 expression with a SUMO mutant resistant to SUMO proteases. Interestingly, we observed that IE1 expression did not affect preSUMOylated PML; however, it clearly prevented de novo SUMO conjugation. Consistent results were obtained by in vitro SUMOylation assays, demonstrating that IE1 alone is sufficient for this effect. Furthermore, IE1 acts in a selective manner, since K160 was identified as the main target lysine. This is strengthened by the fact that IE1 also prevents As2O3-mediated hyperSUMOylation of K160, thereby blocking PML degradation. Since IE1 did not interfere with coiled-coil-mediated PML dimerization, we propose that IE1 affects PML autoSUMOylation either by directly abrogating PML E3 ligase function or by preventing access to SUMO sites. Thus, our data suggest a novel mechanism for how a viral protein counteracts a cellular restriction factor by selectively preventing the de novo SUMOylation at specific lysine residues without affecting global protein SUMOylation.
Importance: The human cytomegalovirus IE1 protein acts as an important antagonist of a cellular restriction mechanism that is mediated by subnuclear structures termed PML nuclear bodies. This function of IE1 is required for efficient viral replication and thus constitutes a potential target for antiviral strategies. In this paper, we further elucidate the molecular mechanism for how IE1 antagonizes PML NBs. We show that tight binding of IE1 to PML interferes with the de novo SUMOylation of a distinct lysine residue that is also the target of stress-mediated hyperSUMOylation of PML. This is of importance since it represents a novel mechanism used by a viral antagonist of intrinsic immunity. Furthermore, it highlights the possibility of developing small molecules that specifically abrogate this PML-antagonistic activity of IE1 and thus inhibit viral replication.
Keywords: PML nuclear bodies; human cytomegalovirus; immediate early 1; intrinsic immunity; nuclear domain 10; sumoylation.
Copyright © 2017 American Society for Microbiology.
Figures



Similar articles
-
The ND10 Component Promyelocytic Leukemia Protein Acts as an E3 Ligase for SUMOylation of the Major Immediate Early Protein IE1 of Human Cytomegalovirus.J Virol. 2017 Apr 28;91(10):e02335-16. doi: 10.1128/JVI.02335-16. Print 2017 May 15. J Virol. 2017. PMID: 28250117 Free PMC article.
-
Expression of Human Cytomegalovirus IE1 Leads to Accumulation of Mono-SUMOylated PML That Is Protected from Degradation by Herpes Simplex Virus 1 ICP0.J Virol. 2018 Nov 12;92(23):e01452-18. doi: 10.1128/JVI.01452-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30258013 Free PMC article.
-
Cytomegalovirus immediate-early 1 proteins form a structurally distinct protein class with adaptations determining cross-species barriers.PLoS Pathog. 2021 Aug 9;17(8):e1009863. doi: 10.1371/journal.ppat.1009863. eCollection 2021 Aug. PLoS Pathog. 2021. PMID: 34370791 Free PMC article.
-
The Human CMV IE1 Protein: An Offender of PML Nuclear Bodies.Adv Anat Embryol Cell Biol. 2017;223:77-94. doi: 10.1007/978-3-319-53168-7_4. Adv Anat Embryol Cell Biol. 2017. PMID: 28528440 Review.
-
Emerging Role of PML Nuclear Bodies in Innate Immune Signaling.J Virol. 2016 Jun 10;90(13):5850-5854. doi: 10.1128/JVI.01979-15. Print 2016 Jul 1. J Virol. 2016. PMID: 27053550 Free PMC article. Review.
Cited by
-
Navigating the Host Cell Response during Entry into Sites of Latent Cytomegalovirus Infection.Pathogens. 2018 Mar 16;7(1):30. doi: 10.3390/pathogens7010030. Pathogens. 2018. PMID: 29547547 Free PMC article. Review.
-
Bright and Early: Inhibiting Human Cytomegalovirus by Targeting Major Immediate-Early Gene Expression or Protein Function.Viruses. 2020 Jan 16;12(1):110. doi: 10.3390/v12010110. Viruses. 2020. PMID: 31963209 Free PMC article. Review.
-
Gammaherpesviral Tegument Proteins, PML-Nuclear Bodies and the Ubiquitin-Proteasome System.Viruses. 2017 Oct 21;9(10):308. doi: 10.3390/v9100308. Viruses. 2017. PMID: 29065450 Free PMC article. Review.
-
Promyelocytic Leukemia Protein Potently Restricts Human Cytomegalovirus Infection in Endothelial Cells.Int J Mol Sci. 2022 Oct 8;23(19):11931. doi: 10.3390/ijms231911931. Int J Mol Sci. 2022. PMID: 36233232 Free PMC article.
-
Cross-Species Analysis of Innate Immune Antagonism by Cytomegalovirus IE1 Protein.Viruses. 2022 Jul 26;14(8):1626. doi: 10.3390/v14081626. Viruses. 2022. PMID: 35893691 Free PMC article.
References
-
- Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF III, Maul GG. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol 147:221–234. doi:10.1083/jcb.147.2.221. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

