Vesivirus 2117 capsids more closely resemble sapovirus and lagovirus particles than other known vesivirus structures
- PMID: 27902397
- PMCID: PMC5370393
- DOI: 10.1099/jgv.0.000658
Vesivirus 2117 capsids more closely resemble sapovirus and lagovirus particles than other known vesivirus structures
Abstract
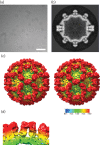
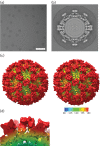
Vesivirus 2117 is an adventitious agent that, in 2009, was identified as a contaminant of Chinese hamster ovary cells propagated in bioreactors at a pharmaceutical manufacturing plant belonging to Genzyme. The consequent interruption in supply of Fabrazyme and Cerezyme (drugs used to treat Fabry and Gaucher diseases, respectively) caused significant economic losses. Vesivirus 2117 is a member of the Caliciviridae, a family of small icosahedral viruses encoding a positive-sense RNA genome. We have used cryo-electron microscopy and three-dimensional image reconstruction to calculate a structure of vesivirus 2117 virus-like particles as well as feline calicivirus and a chimeric sapovirus. We present a structural comparison of several members of the Caliciviridae, showing that the distal P domain of vesivirus 2117 is morphologically distinct from that seen in other known vesivirus structures. Furthermore, at intermediate resolutions, we found a high level of structural similarity between vesivirus 2117 and Caliciviridae from other genera: sapovirus and rabbit hemorrhagic disease virus. Phylogenetic analysis confirms vesivirus 2117 as a vesivirus closely related to canine vesiviruses. We postulate that morphological differences in virion structure seen between vesivirus clades may reflect differences in receptor usage.
Figures




Similar articles
-
Canine caliciviruses of four serotypes from military and research dogs recovered in 1963-1978 belong to two phylogenetic clades in the Vesivirus genus.Virol J. 2018 Feb 23;15(1):39. doi: 10.1186/s12985-018-0944-4. Virol J. 2018. PMID: 29471848 Free PMC article.
-
Caliciviridae Other Than Noroviruses.Viruses. 2019 Mar 21;11(3):286. doi: 10.3390/v11030286. Viruses. 2019. PMID: 30901945 Free PMC article. Review.
-
The Cryo-EM Structure of Vesivirus 2117 Highlights Functional Variations in Entry Pathways for Viruses in Different Clades of the Vesivirus Genus.J Virol. 2021 Jun 10;95(13):e0028221. doi: 10.1128/JVI.00282-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33853966 Free PMC article.
-
Inter- and intragenus structural variations in caliciviruses and their functional implications.J Virol. 2004 Jun;78(12):6469-79. doi: 10.1128/JVI.78.12.6469-6479.2004. J Virol. 2004. PMID: 15163740 Free PMC article.
-
Calicivirus RNA-Dependent RNA Polymerases: Evolution, Structure, Protein Dynamics, and Function.Front Microbiol. 2019 Jun 6;10:1280. doi: 10.3389/fmicb.2019.01280. eCollection 2019. Front Microbiol. 2019. PMID: 31244803 Free PMC article. Review.
Cited by
-
Cryo-Electron Microscopy Structure of the Macrobrachium rosenbergii Nodavirus Capsid at 7 Angstroms Resolution.Sci Rep. 2017 May 18;7(1):2083. doi: 10.1038/s41598-017-02292-0. Sci Rep. 2017. PMID: 28522842 Free PMC article.
-
Canine caliciviruses of four serotypes from military and research dogs recovered in 1963-1978 belong to two phylogenetic clades in the Vesivirus genus.Virol J. 2018 Feb 23;15(1):39. doi: 10.1186/s12985-018-0944-4. Virol J. 2018. PMID: 29471848 Free PMC article.
-
Characterization of a Vesivirus Associated with an Outbreak of Acute Hemorrhagic Gastroenteritis in Domestic Dogs.J Clin Microbiol. 2018 Apr 25;56(5):e01951-17. doi: 10.1128/JCM.01951-17. Print 2018 May. J Clin Microbiol. 2018. PMID: 29444830 Free PMC article.
-
Caliciviridae Other Than Noroviruses.Viruses. 2019 Mar 21;11(3):286. doi: 10.3390/v11030286. Viruses. 2019. PMID: 30901945 Free PMC article. Review.
-
The Cryo-EM Structure of Vesivirus 2117 Highlights Functional Variations in Entry Pathways for Viruses in Different Clades of the Vesivirus Genus.J Virol. 2021 Jun 10;95(13):e0028221. doi: 10.1128/JVI.00282-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33853966 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

